Are Semi-Quantitative Clinical Cultures Inadequate? Comparison to Quantitative Analysis of 1053 Bacterial Isolates from 350 Wounds
Abstract
1. Introduction
2. Materials and Methods
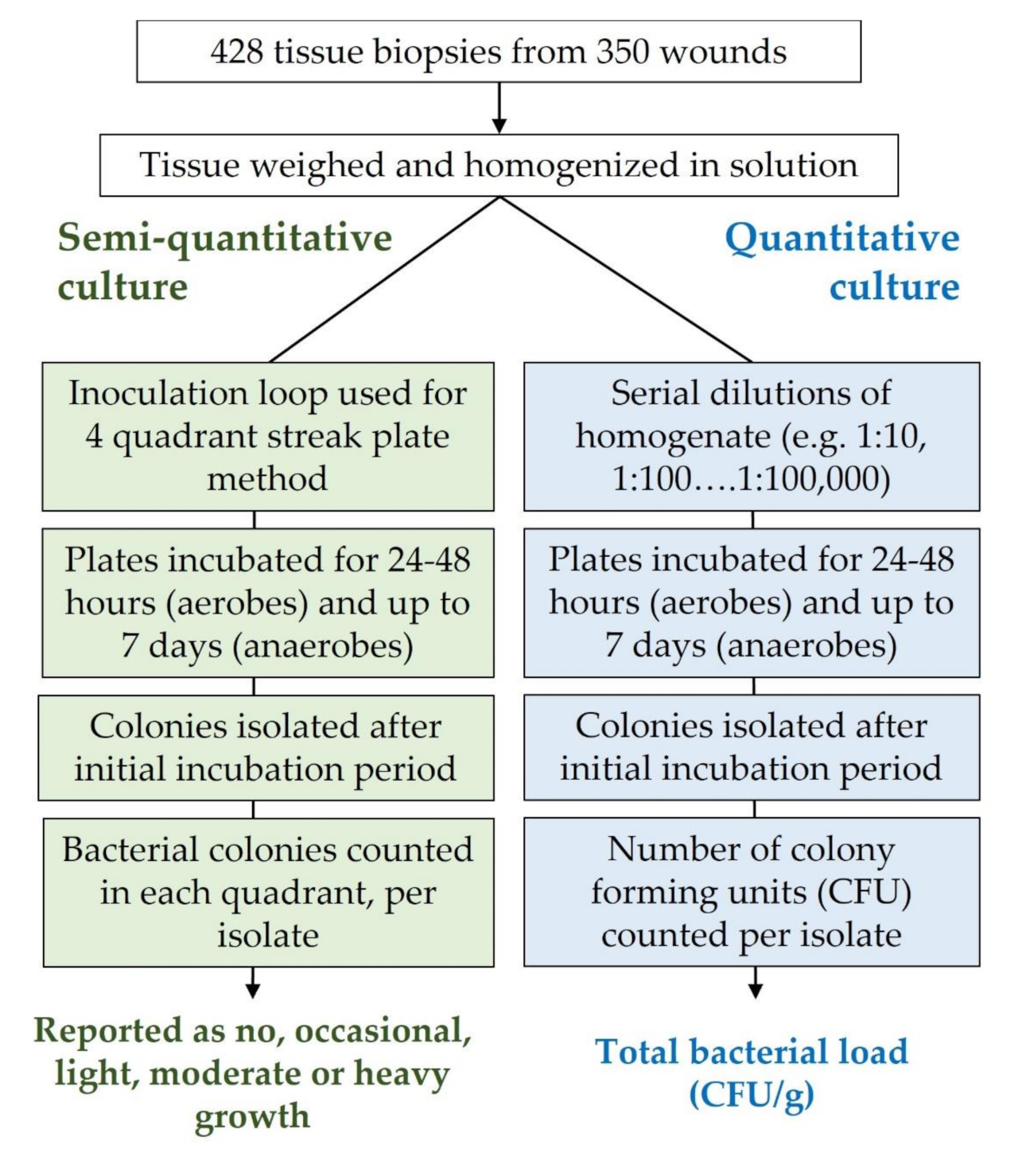
2.1. Wound Biopsy, Semi-Quantitative and Quantitative Culture Analysis
2.2. Fluorescence Imaging Procedure
3. Results
4. Discussion
Proposed Solution
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lantis, J.C., 2nd; Marston, W.A.; Farber, A.; Kirsner, R.S.; Zhang, Y.; Lee, T.D.; Cargill, D.I.; Slade, H.B. The influence of patient and wound variables on healing of venous leg ulcers in a randomized controlled trial of growth-arrested allogeneic keratinocytes and fibroblasts. J. Vasc. Surg. 2013, 58, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S.B.; Shruptha, P.; Prabhu, V.; Srujan, C.; Nayak, U.Y.; Anuradha, C.K.R.; Ramachandra, L.; Keerthana, P.; Joshi, M.B.; Murali, T.S.; et al. Pseudomonas aeruginosa virulence proteins pseudolysin and protease IV impede cutaneous wound healing. Lab. Investig. 2020, 100, 1532–1550. [Google Scholar] [CrossRef]
- Gardner, S.E.; Frantz, R.A. Wound bioburden and infection-related complications in diabetic foot ulcers. Biol. Res. Nurs. 2008, 10, 44–53. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Snyder, R.J.; Bohn, G.; Hanft, J.; Harkless, L.; Kim, P.; Lavery, L.; Schultz, G.; Wolcott, R. Wound Biofilm: Current Perspectives and Strategies on Biofilm Disruption and Treatments. Wounds 2017, 29, S1–S17. [Google Scholar]
- Metcalf, D.G.; Bowler, P.G. Biofilm delays wound healing: A review of the evidence. Burns Trauma 2013, 1, 5–12. [Google Scholar] [CrossRef]
- Caldwell, M.D. Bacteria and Antibiotics in Wound Healing. Surg. Clin. 2020, 100, 757–776. [Google Scholar] [CrossRef]
- Xu, L.; McLennan, S.V.; Lo, L.; Natfaji, A.; Bolton, T.; Liu, Y.; Twigg, S.M.; Yue, D.K. Bacterial load predicts healing rate in neuropathic diabetic foot ulcers. Diabetes Care 2007, 30, 378–380. [Google Scholar] [CrossRef] [PubMed]
- Majewski, W.; Cybulski, Z.; Napierala, M.; Pukacki, F.; Staniszewski, R.; Pietkiewicz, K.; Zapalski, S. The value of quantitative bacteriological investigations in the monitoring of treatment of ischaemic ulcerations of lower legs. Int. Angiol. 1995, 14, 381–384. [Google Scholar] [PubMed]
- Lookingbill, D.P.; Miller, S.H.; Knowles, R.C. Bacteriology of chronic leg ulcers. Arch. Dermatol. 1978, 114, 1765–1768. [Google Scholar] [CrossRef]
- Robson, M.C.; Heggers, J.P. Delayed wound closure based on bacterial counts. J. Surg. Oncol. 1970, 2, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Woodmansey, E.J.; Roberts, C.D. Appropriate use of dressings containing nanocrystalline silver to support antimicrobial stewardship in wounds. Int. Wound J. 2018, 15, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.E.; Frantz, R.A.; Doebbeling, B.N. The validity of the clinical signs and symptoms used to identify localized chronic wound infection. Wound Repair Regen. 2001, 9, 178–186. [Google Scholar] [CrossRef]
- Gardner, S.E.; Hillis, S.L.; Frantz, R.A. Clinical signs of infection in diabetic foot ulcers with high microbial load. Biol. Res. Nurs. 2009, 11, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Le, L.; Baer, M.; Briggs, P.; Bullock, N.; Cole, W.; DiMarco, D.; Hamil, R.; Harrell, K.; Kasper, M.; Li, W.; et al. Diagnostic Accuracy of Point-of-Care Fluorescence Imaging for the Detection of Bacterial Burden in Wounds: Results from the 350-Patient Fluorescence Imaging Assessment and Guidance Trial. Adv. Wound Care 2021, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Serena, T.; Robson, M.C.; Cooper, D.M.; Ignatius, J. Lack of Reliability of Clinical/Visual Assessment of Chronic Wound Infection: The Incidence of Biopsy-Proven Infection in Venous Leg Ulcers. Wounds 2006, 18, 5. [Google Scholar]
- Reddy, M.; Gill, S.S.; Wu, W.; Kalkar, S.R.; Rochon, P.A. Does this patient have an infection of a chronic wound? JAMA 2012, 307, 605–611. [Google Scholar] [CrossRef]
- Raizman, R.; Little, W.; Smith, A.C. Rapid Diagnosis of Pseudomonas aeruginosa in Wounds with Point-Of-Care Fluorescence Imaing. Diagnostics 2021, 11, 280. [Google Scholar] [CrossRef]
- Davies, J.A.; Bull, R.H.; Farrelly, I.J.; Wakelin, M.J. A home-based exercise programme improves ankle range of motion in long-term venous ulcer patients. Phlebology 2007, 22, 86–89. [Google Scholar] [CrossRef]
- Dow, G. Bacterial swabs and the chronic wound: When, how, and what do they mean? Ostomy Wound Manag. 2003, 49, 8–13. [Google Scholar]
- Angel, D.E.; Lloyd, P.; Carville, K.; Santamaria, N. The clinical efficacy of two semi-quantitative wound-swabbing techniques in identifying the causative organism(s) in infected cutaneous wounds. Int. Wound. J. 2011, 8, 176–185. [Google Scholar] [CrossRef]
- Buchanan, R.B.; Blamey, R.W.; Durrant, K.R.; Howell, A.; Paterson, A.G.; Preece, P.E.; Smith, D.C.; Williams, C.J.; Wilson, R.G. A randomized comparison of tamoxifen with surgical oophorectomy in premenopausal patients with advanced breast cancer. J. Clin. Oncol. 1986, 4, 1326–1330. [Google Scholar] [CrossRef]
- Buchanan, K.; Heimbach, D.M.; Minshew, B.H.; Coyle, M.B. Comparison of quantitative and semiquantitative culture techniques for burn biopsy. J. Clin. Microbiol. 1986, 23, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Thomson, P.D.; Smith, D.J. Jr. What Is Infection? Am. J. Sur. 1994, 167, S7–S11. [Google Scholar] [CrossRef]
- Gardner, S.E.; Frantz, R.A.; Park, H.; Scherubel, M. The inter-rater reliability of the Clinical Signs and Symptoms Checklist in diabetic foot ulcers. Ostomy Wound Manag. 2007, 53, 46–51. [Google Scholar]
- Ratliff, C.R.; Rodeheaver, G.T. Correlation of semi-quantitative swab cultures to quantitative swab cultures from chronic wounds. Wounds 2002, 14, 329–333. [Google Scholar]
- Sauget, M.; Valot, B.; Bertrand, X.; Hocquet, D. Can MALDI-TOF Mass Spectrometry Reasonably Type Bacteria? Trends in Microbiol. 2017, 25, 447–455. [Google Scholar] [CrossRef]
- Jones, L.M.; Dunham, D.; Rennie, M.Y.; Kirman, J.; Lopez, A.J.; Keim, K.C.; Little, W.; Gomez, A.; Bourke, J.; Ng, H.; et al. In vitro detection of porphyrin-producing wound bacteria with real-time fluorescence imaging. Future Microbiol. 2020, 15, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.Y.; Dunham, D.; Lindvere-Teene, L.; Raizman, R.; Hill, R.; Linden, R. Understanding real-time fluorescence signals from Bacteria and wound tissues observed with the MolecuLight i:X(TM). Diagnostics 2019, 9, 22. [Google Scholar] [CrossRef]
- Serena, T.E.; Harrell, K.; Serena, L.; Yaakov, R.A. Real-time bacterial fluorescence imaging accurately identifies wounds with moderate-to-heavy bacterial burden. J. Wound Care 2019, 28, 346–357. [Google Scholar] [CrossRef]
- Gardner, S.E.; Frantz, R.; Hillis, S.L.; Park, H.; Scherubel, M. Diagnostic Validity of Semiquantitative Swab Cultures. Wounds 2007, 19, 31–38. [Google Scholar]
- Thomsen, T.R.; Aasholm, M.S.; Rudkjobing, V.B.; Saunders, A.M.; Bjarnsholt, T.; Givskov, M.; Kirketerp-Moller, K.; Nielsen, P.H. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Repair Regen. 2010, 18, 38–49. [Google Scholar] [CrossRef]
- Cutting, K. Wound infection conundrum. Br. J. Nurs. 2013, 22, S3. [Google Scholar] [CrossRef] [PubMed]
- Lipsky, B.A.; Dryden, M.; Gottrup, F.; Nathwani, D.; Seaton, R.A.; Stryja, J. Antimicrobial stewardship in wound care: A position paper from the British society for Antimicrobial Chemotherapy and European Wound Management Association. J. Antimicrob. Chemother. 2016, 71, 3026–3035. [Google Scholar] [CrossRef]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global Contributors to Antibiotic Resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Action Plan on Antimicrobial Resistance. 2015. Available online: https://www.who.int/iris/handle/10665/193736 (accessed on 31 May 2021).
- Bernier, A.; Delarocque-Astagneau, E.; Ligier, C.; Vibet, M.-A.; Guillemot, D.; Watier, L. Outpatient Antibiotic Use in France between 2000 and 2010: After the Nationwide Campaign, It Is Time To Focus on the Elderly. Antimicrob. Agents Chemother. 2014, 58, 71–77. [Google Scholar] [CrossRef][Green Version]
- Mattappalil, A.; Mergenhagen, K.A. Neurotoxicity with antimicrobials in the elderly: A review. Clin. Ther. 2014, 36, 1489–1511.e4. [Google Scholar] [CrossRef]
- Oliver, J.D. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef]
- Zandri, G.; Pasquaroli, S.; Vignaroli, C.; Talevi, S.; Manso, E.; Donelli, G.; Biavasco, F. Detection of viable but non-culturable staphylococci in biofilms from central venous catheters negative on standard microbiological assays. Clin. Microbiol. Infect. 2012, 18, E259–E261. [Google Scholar] [CrossRef]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016, 24, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.D.; Wolcott, R.D.; Sun, Y.; Dowd, S.E. Comparison of culture and molecular identification of bacteria in chronic wounds. Int. J. Mol. Sci. 2012, 13, 2535–2550. [Google Scholar] [CrossRef]
- Serena, T.E.; Bayliff, S.W.; Brosnan, P.J.; DiMarco, D.T.; Doner, B.A.; Guthrie, D.A.; Patel, K.D.; Sabo, M.J.; Samies, J.H.; Carter, M.J. Bacterial protease activity as a biomarker to assess the risk of non-healing in chronic wounds: Results from a multicentre randomised controlled clinical trial. Wound Repair Regen. 2021. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, G.; Schultz, G.; Kitamura, A.; Minematsu, T.; Akamata, K.; Suga, H.; Kurita, M.; Hayashi, C.; Sanada, H. Rapid detection of biofilm by wound blotting following sharp debridement of chronic pressure ulcers predicts wound healing: A preliminary study. Int. Wound J. 2020, 17, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.J.; Jones, L.M.; Reynolds, L.; Diaz, R.C.; George, I.K.; Little, W.; Fleming, D.; D’Souza, A.; Rennie, M.Y.; Rumbaugh, K.P.; et al. Detection of bacterial fluorescence from in vivo wound biofilms using a point-of-care fluorescence imaging device. Int. Wound J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Cole, W.; Coe, S. Use of a bacterial fluorescence imaging system to target wound debridement and accelerate healing: A pilot study. J. Wound Care 2020, 29, S44–S52. [Google Scholar] [CrossRef]
- Hill, R.; Woo, K. A prospective multisite observational study incorporating bacterial fluorescence information into the upper/lower wound infection checklists. Wounds 2020, 32, 299–308. [Google Scholar] [PubMed]
- Raizman, R.; Dunham, D.; Lindvere-Teene, L.; Jones, L.M.; Tapang, K.; Linden, R.; Rennie, M.Y. Use of a bacterial fluorescence imaging device: Wound measurement, bacterial detection and targeted debridement. J. Wound Care 2019, 28, 824–834. [Google Scholar] [CrossRef]
- Price, N. Routine fluorescence imaging to detect wound bacteria reduces antibiotic use and antimicrobial dressing expenditure while improving healing rates: Retrospective analysis of 229 foot ulcers. Diagnostics 2020, 10, 927. [Google Scholar] [CrossRef]
- Serena, T.E. Incorporating point-of-care bacterial fluorescence into a wound clinic antimicrobial stewardship program. Diagnostics 2020, 10, 1010. [Google Scholar] [CrossRef]

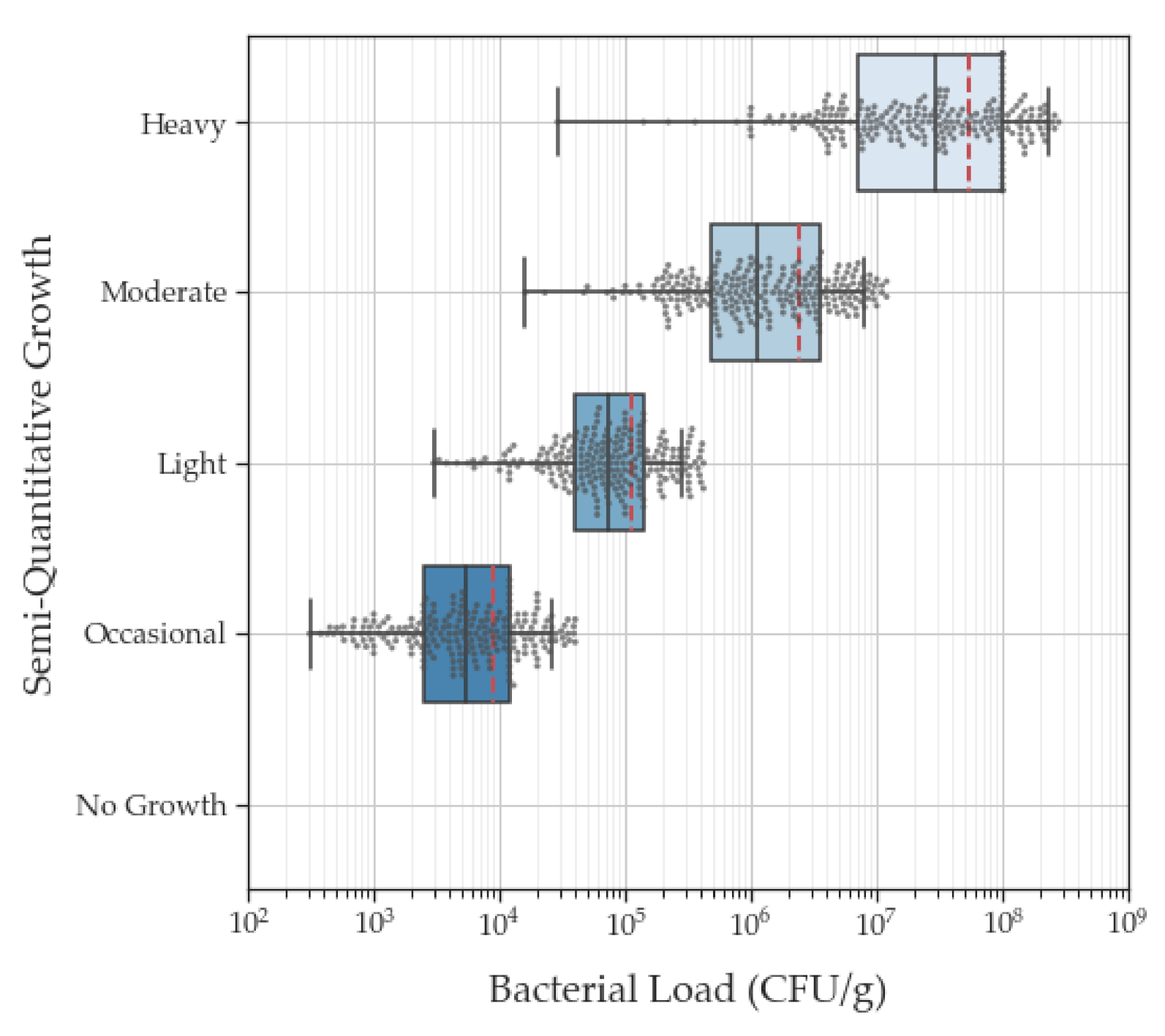

| Semi-Quantitative Culture Categories | Quantitative Culture Values | |
|---|---|---|
| Mean (SE) | Median (Range) | |
| Occasional | 4.9 × 104 CFU/g (3.1 × 104 CFU/g) | 6.6 × 103 CFU/g (3.1 × 102 CFU/g–7.3 × 106 CFU/g) |
| Light | 2.5 × 105 CFU/g (6.3 × 104 CFU/g) | 8.6 × 104 CFU/g (3.0 × 103–1.4 × 107 CFU/g) |
| Moderate | 5.4 × 106 CFU/g (6.1 × 105 CFU/g) | 1.6 × 106 CFU/g (1.6 × 104–7.3 × 107 CFU/g) |
| Heavy | 1.4 × 108 CFU/g (2.9 × 107 CFU/g) | 3.5 × 107 CFU/g (2.9 × 104–7.0 × 109 CFU/g) |
| Semi-Quantitative Category | ||||
|---|---|---|---|---|
| Quantitative Bacterial Load | Occasional N = 238 | Light N = 246 | Moderate N = 272 | Heavy N = 257 |
| <104 CFU/g | 142 | 10 | 0 | 0 |
| 104 CFU/g | 88 | 127 | 8 | 1 |
| 105 CFU/g | 7 | 103 | 99 | 7 |
| 106 CFU/g | 1 | 5 | 128 | 64 |
| 107 CFU/g | 0 | 1 | 37 | 97 |
| >108 CFU/g | 0 | 0 | 0 | 88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serena, T.E.; Bowler, P.G.; Schultz, G.S.; D’souza, A.; Rennie, M.Y. Are Semi-Quantitative Clinical Cultures Inadequate? Comparison to Quantitative Analysis of 1053 Bacterial Isolates from 350 Wounds. Diagnostics 2021, 11, 1239. https://doi.org/10.3390/diagnostics11071239
Serena TE, Bowler PG, Schultz GS, D’souza A, Rennie MY. Are Semi-Quantitative Clinical Cultures Inadequate? Comparison to Quantitative Analysis of 1053 Bacterial Isolates from 350 Wounds. Diagnostics. 2021; 11(7):1239. https://doi.org/10.3390/diagnostics11071239
Chicago/Turabian StyleSerena, Thomas E., Philip G. Bowler, Gregory S. Schultz, Anna D’souza, and Monique Y. Rennie. 2021. "Are Semi-Quantitative Clinical Cultures Inadequate? Comparison to Quantitative Analysis of 1053 Bacterial Isolates from 350 Wounds" Diagnostics 11, no. 7: 1239. https://doi.org/10.3390/diagnostics11071239
APA StyleSerena, T. E., Bowler, P. G., Schultz, G. S., D’souza, A., & Rennie, M. Y. (2021). Are Semi-Quantitative Clinical Cultures Inadequate? Comparison to Quantitative Analysis of 1053 Bacterial Isolates from 350 Wounds. Diagnostics, 11(7), 1239. https://doi.org/10.3390/diagnostics11071239







