Gamma-Glutamyl Transferase (GGT) Is the Leading External Quality Assurance Predictor of ISO15189 Compliance for Pathology Laboratories
Abstract
1. Introduction
2. Methods
2.1. Data
2.1.1. NATA Results and Response Classes
2.1.2. Calculation of Relative Deviation for EQA Markers
2.1.3. Direct Comparison of EQA and NATA Results
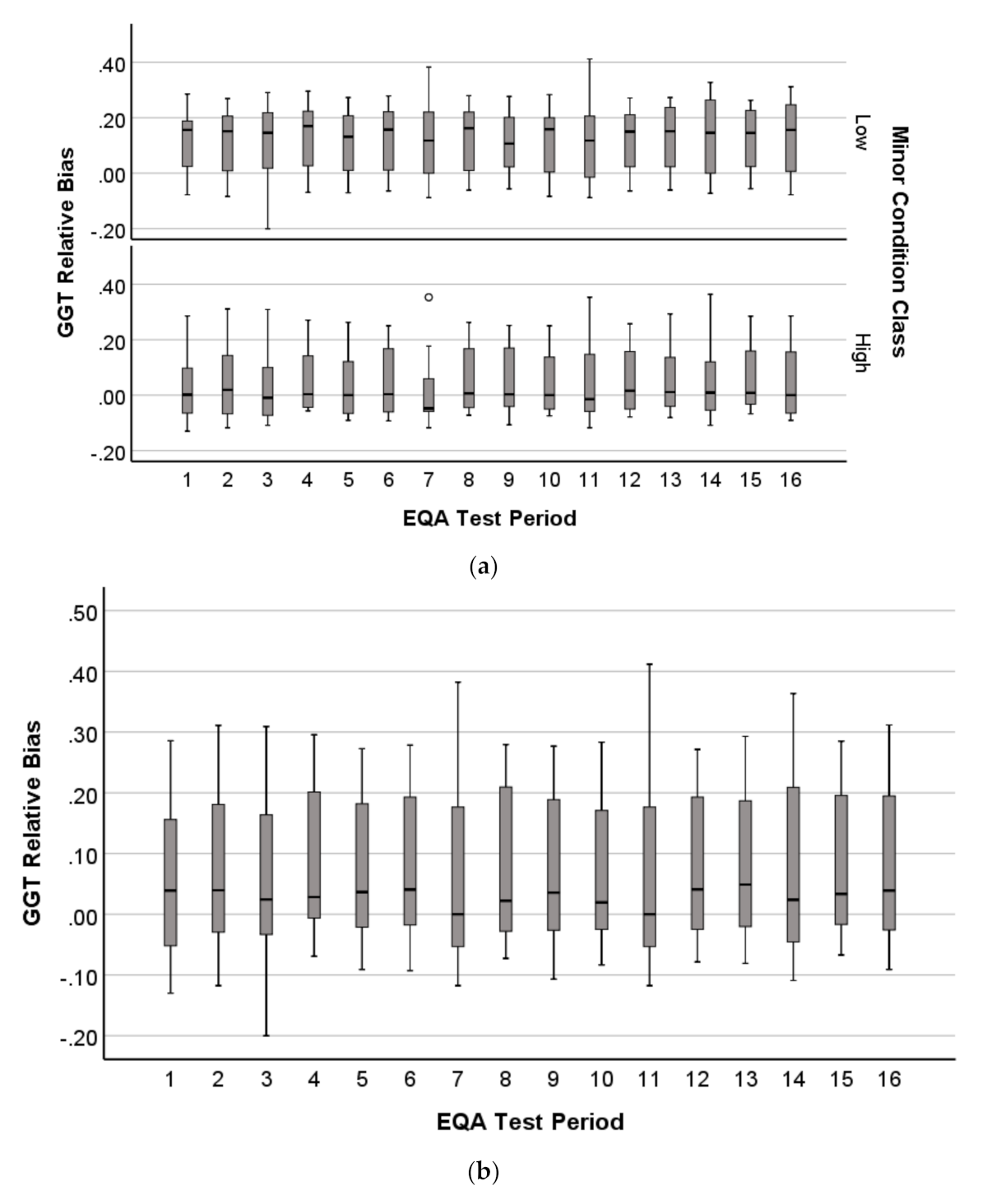
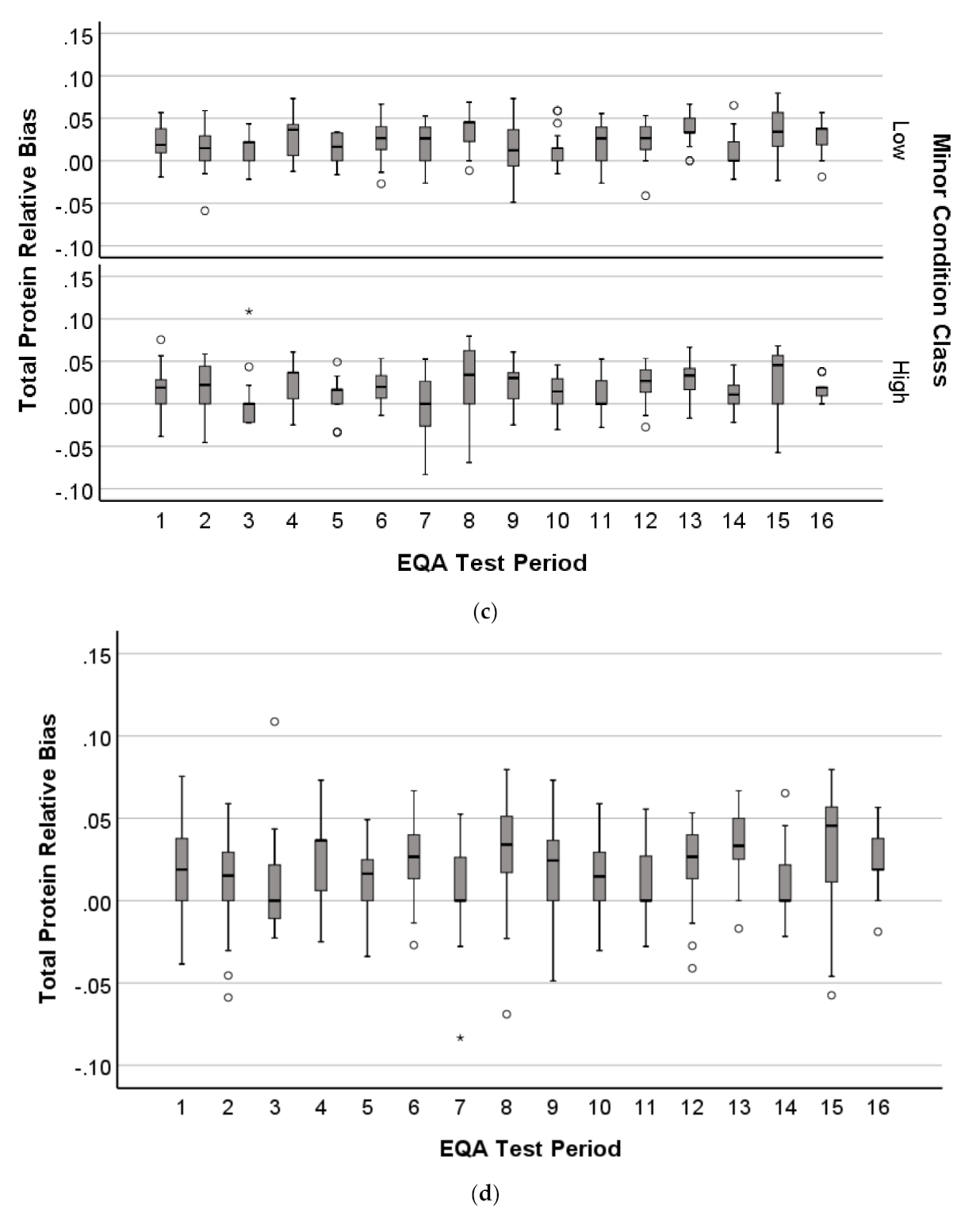
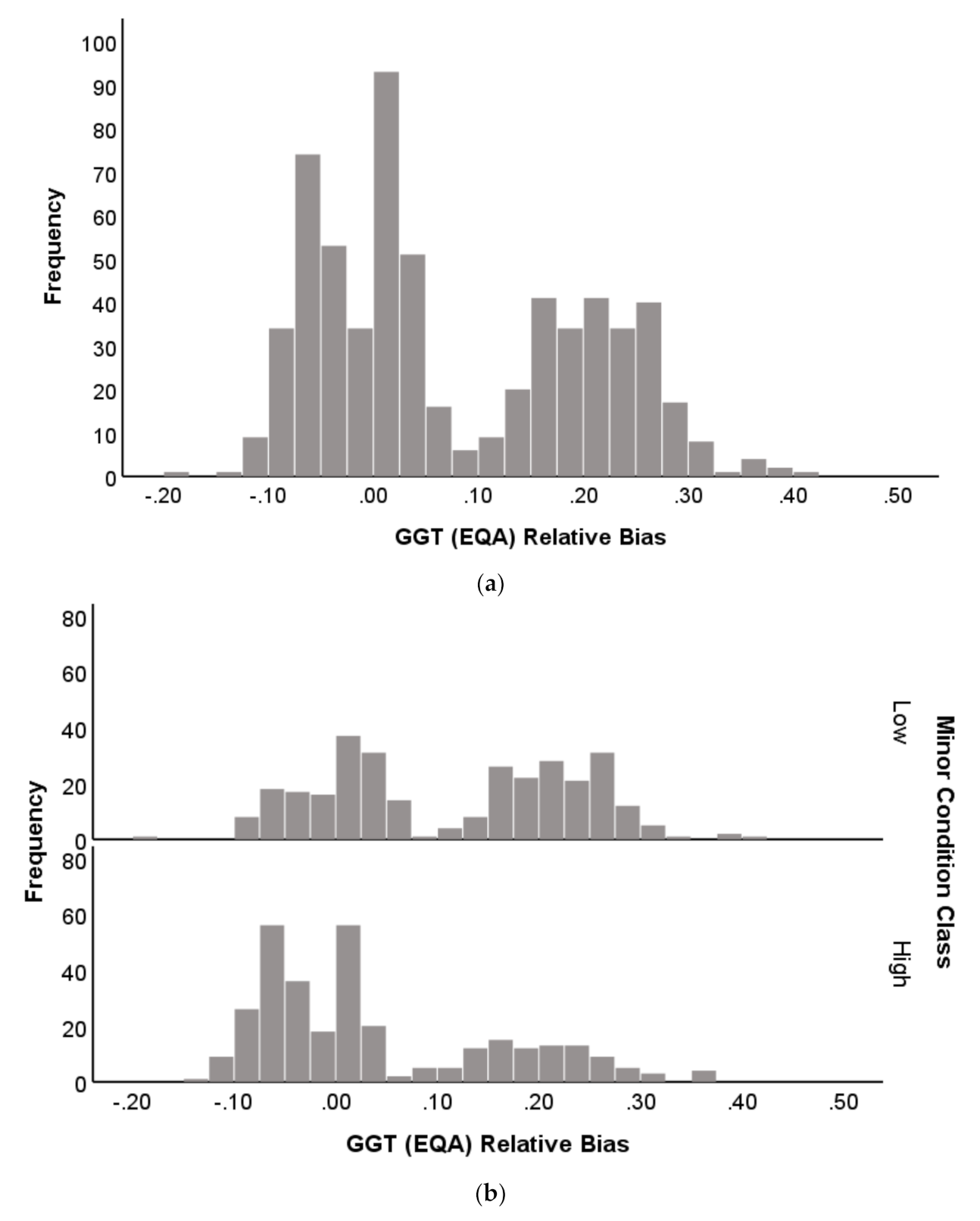
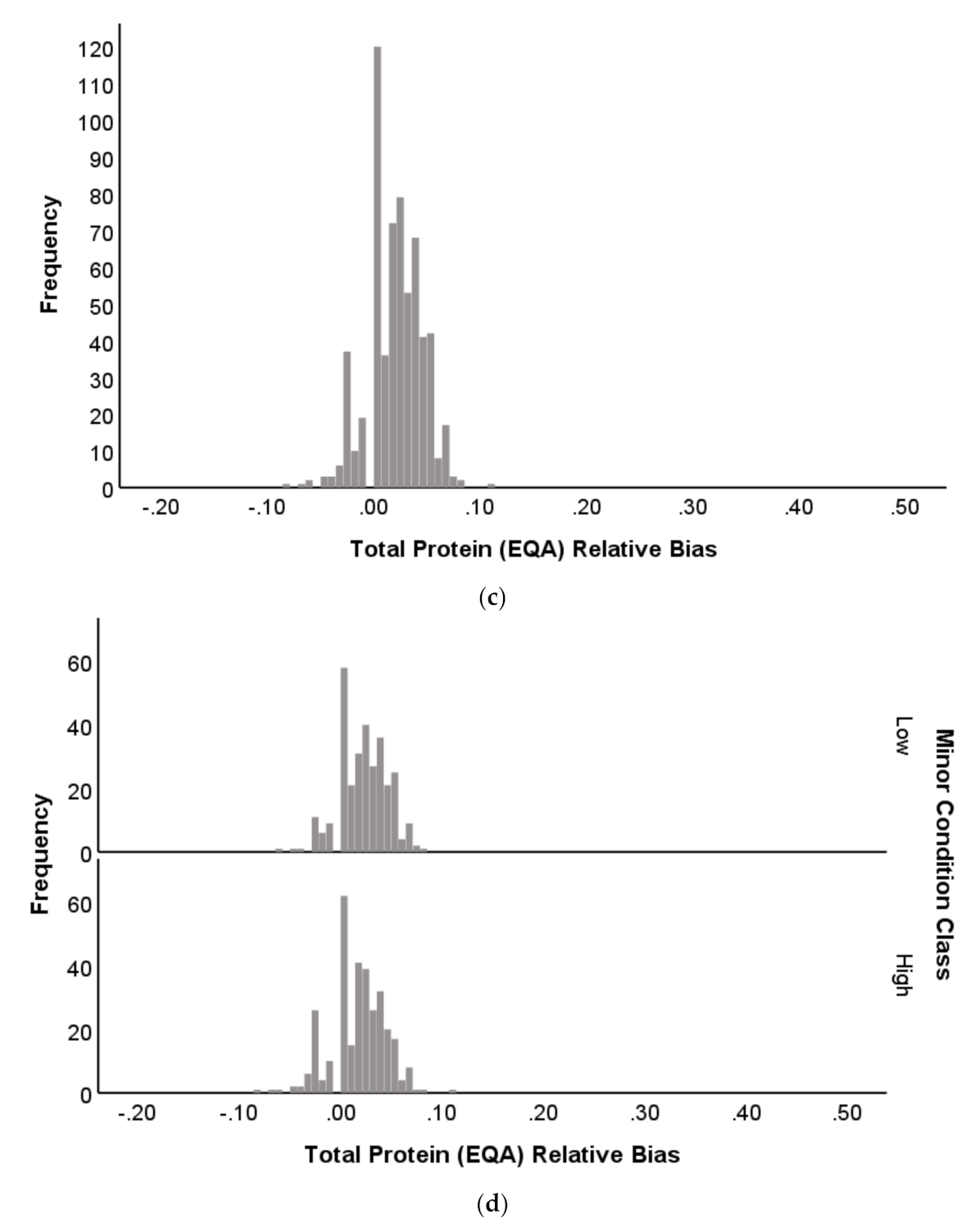
2.1.4. Distribution Profile of EQA Performance
2.2. Statistical Methods
2.3. Machine Learning-Integration of NATA and EQA Results
3. Results
3.1. Direct Relationship of EQA Performance to NATA (ISO15189) Compliance
3.2. Arrangement of EQA Results in Relation to NATA Response
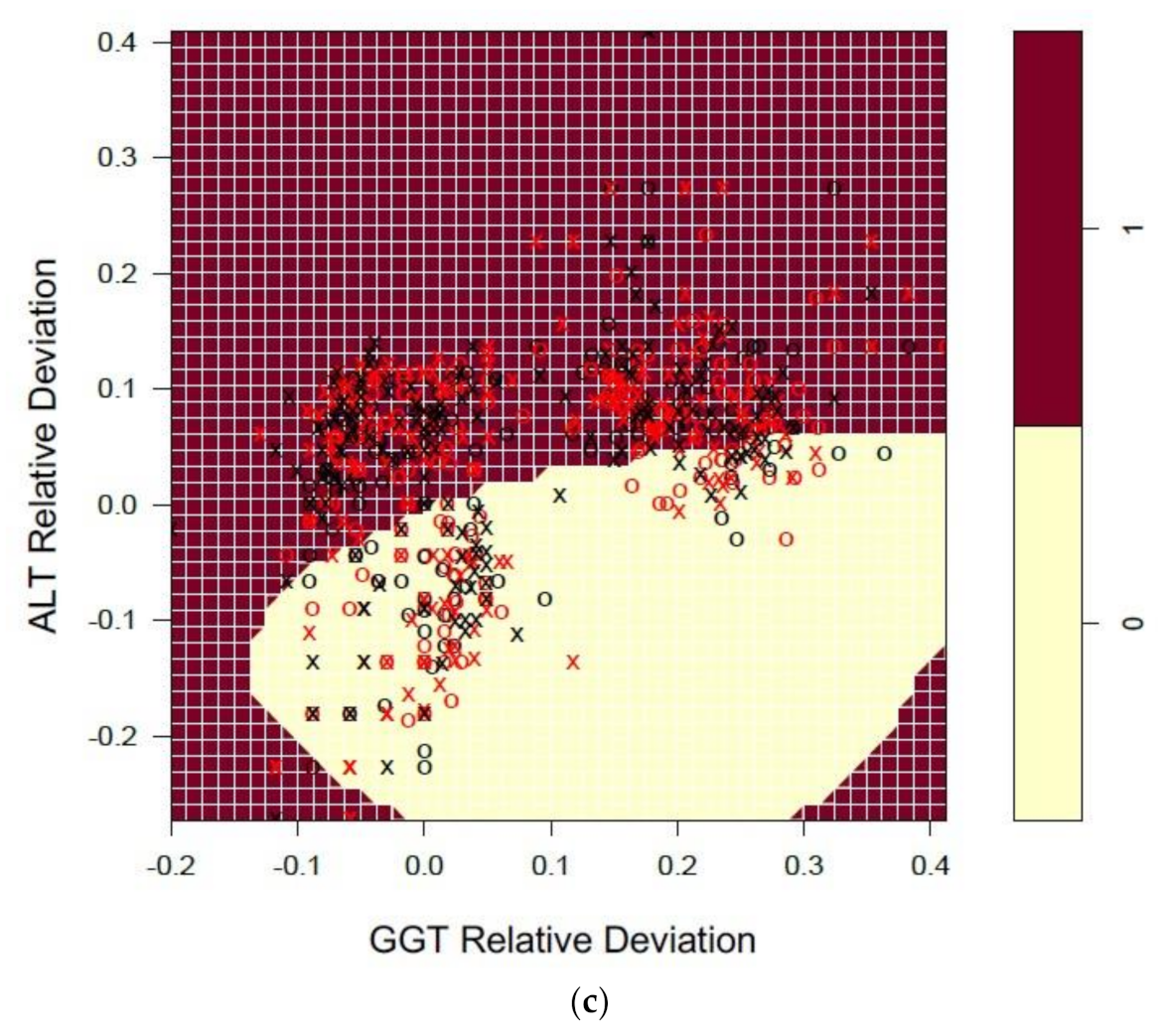
3.3. Machine Learning-Predicting NATA Compliance by EQA Relative Deviation
3.3.1. Creatinine and Electrolytes
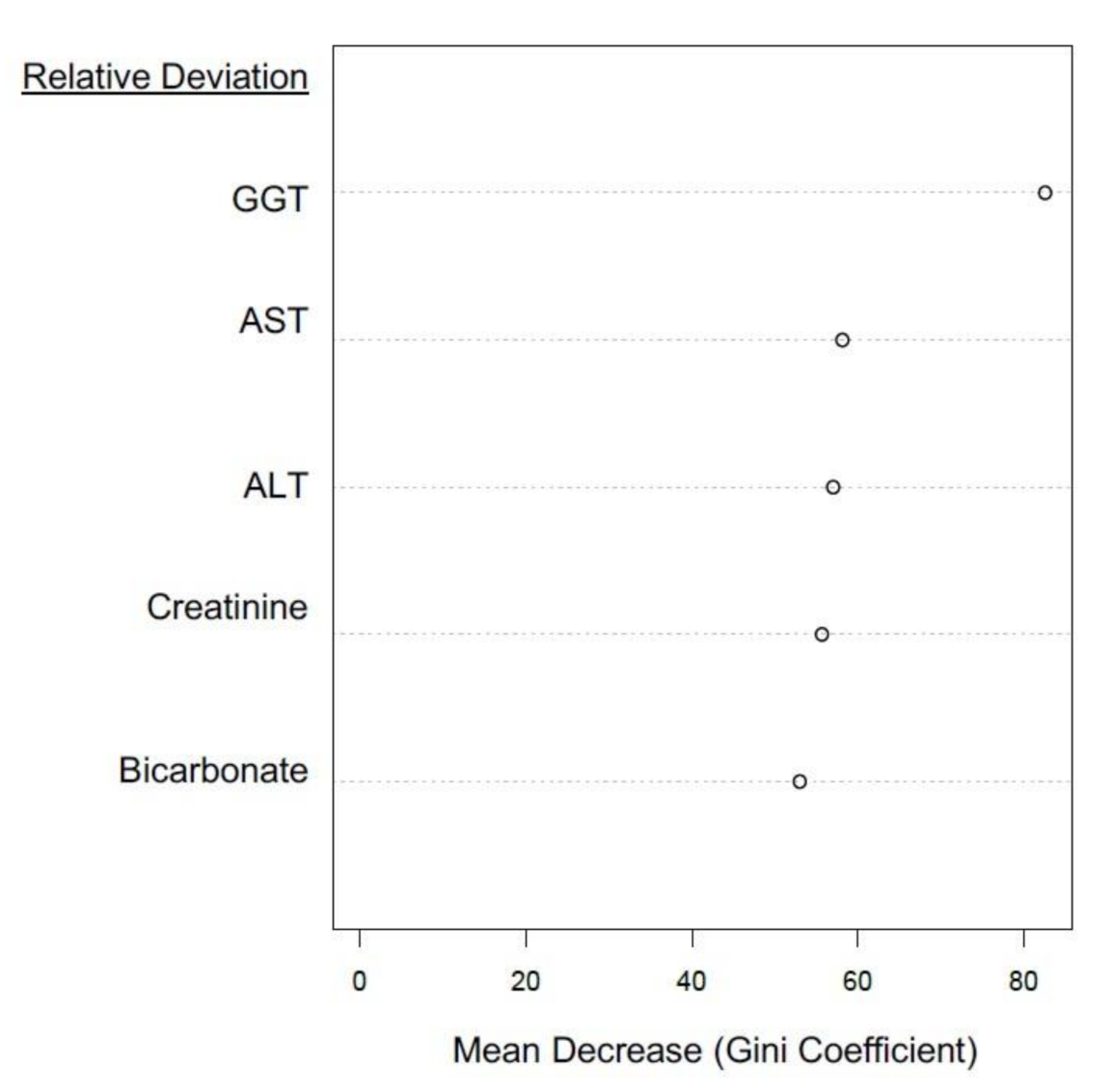
3.3.2. Liver Function Tests (LFTs)
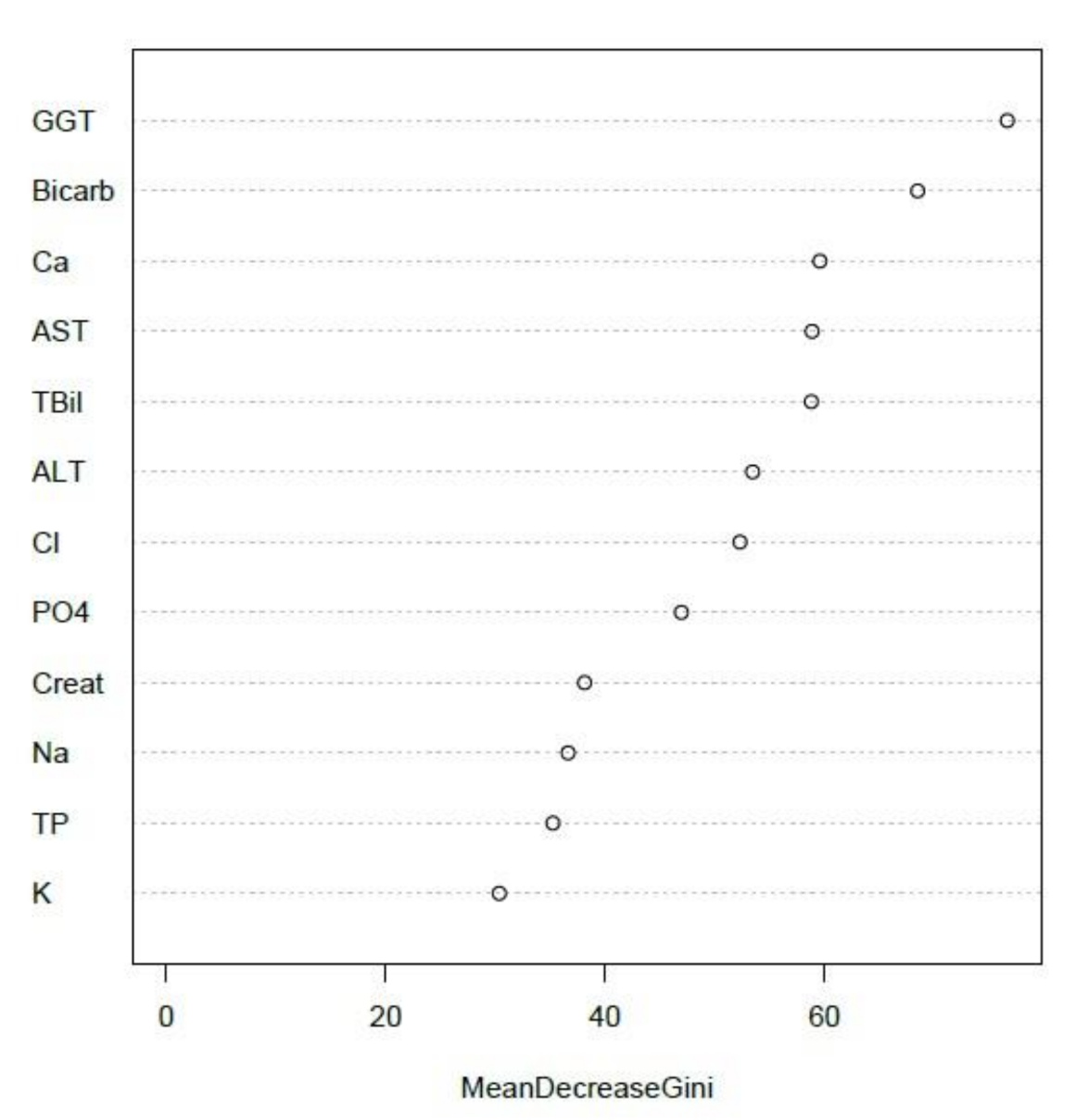
3.3.3. NATA Class Prediction by Combined Creatinine-Electrolyte-LFT EQA Results
3.3.4. Investigations of EQA Marker Predictive Capacity via Unsupervised Analyses
3.4. Patterns in EQA Relative Deviation Distribution and the Relationship to NATA Class Prediction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sciacovelli, L.; Aita, A.; Padoan, A.; Antonelli, G.; Plebani, M. ISO 15189 accreditation and competence: A new opportunity for laboratory medicine. J. Lab. Precis. Med. 2017, 2, 79. [Google Scholar] [CrossRef]
- Gay, S.; Badrick, T.; Ross, J. “State of the art” for competency assessment in Australian medical laboratories. Accredit. Qual. Assur. 2020, 25, 323–327. [Google Scholar] [CrossRef]
- Plebani, M. Quality in laboratory medicine: An unfinished journey. J. Lab. Precis. Med. 2017, 2, 63. [Google Scholar] [CrossRef]
- Badrick, T.; Bietenbeck, A.; Cervinski, M.; Katayev, A.; van Rossum, H.; Loh, T. International Federation of Clinical Chemistry, and Laboratory Medicine Committee on Analytical Quality. Patient-based real-time quality control: Review and recommendations. Clin. Chem. 2019, 65, 962–971. [Google Scholar] [CrossRef]
- NATA. National Association of Testing Authorities, Australia. 2017. Available online: www.nata.com.au/nata/ (accessed on 11 August 2020).
- Lidbury, B.A.; Koerbin, G.; Richardson, A.M.; Badrick, T. Integration of ISO 15189 and external quality assurance data to assist the detection of poor laboratory performance in NSW, Australia. J. Lab. Precis. Med. 2017, 2, 97. [Google Scholar] [CrossRef]
- National Pathology Accreditation Advisory Council (NPAAC). The Requirements for Supervision in the Clinical Governance of Medical Pathology Laboratories (Fourth Edition 2018). Available online: https://www1.health.gov.au/internet/main/Publishing.nsf/Content/74F0211140E493B9CA257BF0001FEAB2/$File/20180612-Final-Supervison-in-the-Clinical-Governance.pdf (accessed on 3 September 2020).
- IBM Corp. IBM SPSS Statistics for Windows; IBM Corp: Armonk, NY, USA, 2019. [Google Scholar]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 11 August 2020).
- Karatzoglou, A.; Meyer, D.; Hornik, K. Support Vector Machines in R. J. Stat. Softw. 2006, 15, 1–28. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R Package Version. 2019. Available online: https://CRAN.R-project.org/package=e1071 (accessed on 11 August 2020).
- Therneau, T.; Atkinson, B. rpart: Recursive Partitioning and Regression Trees. R Package Version 4.1-15. 2019. Available online: https://CRAN.R-project.org/package=rpart (accessed on 11 August 2020).
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Caret: Classification and Regression Training. R Package Version 6.0-84. 2019. Available online: https://CRAN.R-project.org/package=caret (accessed on 11 August 2020).
- White, B. The impact of ISO 15189 and ISO 9001 quality management systems on reducing errors. Vox Sang 2002, 83 (Suppl. 1), 17–20. [Google Scholar] [CrossRef]
- Hartley, T.F. An audit questionnaire that examines specifically the management of technical activities clauses in ISO 15189. Indian J. Clin. Biochem. 2010, 25, 92–98. [Google Scholar] [CrossRef]
- Ho, B.; Ho, E. The most common nonconformities encountered during the assessments of medical laboratories in Hong Kong using ISO 15189 as accreditation criteria. Biochem. Med. 2012, 22, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Farmer, T. Toward a culture shift in laboratory quality: Application of the full ISO 15189 standard. MLO Med Lab. Obs. 2015, 47, 38–39. [Google Scholar] [PubMed]
- Plebani, M.; Sciacovelli, L.; Chiozza, M.L.; Panteghini, M. Once upon a time: A tale of ISO15189 accreditation. Clin. Chem. Lab. Med. 2015, 53, 1127–1129. [Google Scholar] [CrossRef]
- Thelen, M.H.; Vanstapel, F.J.; Kroupis, C.; Vukasovic, I.; Boursier, G.; Barrett, E.; Huisman, W. Flexible scope for ISO15189 accreditation: A guidance prepared by the European Federation of Clinical Chemistry and Laboratory Medicine (EFLM) Working Group Accreditation and ISO/CEN standards (WG-A/ISO). Clin. Chem. Lab. Med. 2015, 53, 1173–1180. [Google Scholar] [CrossRef]
- Gay, S.; Badrick, T. Changes in error rates in the Australian key incident monitoring and management system program. Biochem. Medica 2020, 30, 257–264. [Google Scholar] [CrossRef]
- Loh, T.P.; Badrick, T. Using next generation electronic medical records for laboratory quality monitoring. J. Lab. Precis. Med. 2017, 2, 61. [Google Scholar] [CrossRef]
- Bellini, C.; Cinci, F.; Scapellato, C.; Guerranti, R. A computer model for professional competence assessment according to ISO 15189. Clin. Chem. Lab. Med. 2020, 58, 1242–1249. [Google Scholar] [CrossRef]
- Habli, I.; Lawton, T.; Porter, Z. Artificial intelligence in health care: Accountability and safety. Bull. World Heal. Organ. 2020, 98, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Crowley, S.; Tognarini, D.; Desmond, P.; Lees, M.; Saal, G. Introduction of lamivudine for the treatment of chronic hepatitis B: Expected clinical and economic outcomes based on 4-year clinical trial data. J. Gastroenterol. Hepatol. 2002, 17, 153–164. [Google Scholar] [CrossRef]
- Shang, G.; Richardson, A.; Gahan, M.E.; Easteal, S.; Ohms, S.; Lidbury, B.A. Predicting the presence of hepatitis B virus surface antigen in Chinese patients by pathology data mining. J. Med. Virol. 2013, 85, 1334–1339. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.M.; Lidbury, B.A. Enhancement of hepatitis virus immunoassay outcome predictions in imbalanced routine pathology data by data balancing and feature selection before the application of support vector machines. BMC Med. Inform. Decis. Mak. 2017, 17, 121. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.M.; Lidbury, B.A. Infection status outcome, machine learning method and virus type interact to affect the optimised prediction of hepatitis virus immunoassay results from routine pathology laboratory assays in unbalanced data. BMC Bioinform. 2013, 14, 206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lidbury, B.A.; Richardson, A.M.; Badrick, T. Assessment of machine-learning techniques on large pathology data sets to address assay redundancy in routine liver function test profiles. Diagnosis 2015, 2, 41–51. [Google Scholar] [CrossRef]
- Abels, E.; Pantanowitz, L.; Aeffner, F.; Zarella, M.D.; van der Laak, J.; Bui, M.M.; Agosto-Arroyo, E. Computational pathology definitions, best practices, and recommendations for regulatory guidance: A white paper from the Digital Pathology Association. J. Pathol. 2019, 249, 286–294. [Google Scholar] [CrossRef]
- Williams, B.J.; Knowles, C.; Treanor, D. Maintaining quality diagnosis with digital pathology: A practical guide to ISO 15189 accreditation. J. Clin. Pathol. 2019, 72, 663–668. [Google Scholar] [CrossRef]
- Van Es, S.L. Digital pathology: Semper ad meliora. Pathology 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Volynskaya, Z.; Chow, H.; Evans, A.; Wolff, A.; Lagmay-Traya, C.; Asa, S.L. Integrated pathology informatics enables high-quality personalized and precision medicine: Digital pathology and beyond. Arch. Pathol. Lab. Med. 2018, 142, 369–382. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Olshen, R.; Stone, C. Classification and Regression Trees; Wadsworth International Group: Belmont, CA, USA, 1984. [Google Scholar]
- Kingsford, C.; Salzberg, S.L. What are decision trees? Nat. Biotechnol. 2008, 26, 1011–1013. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]

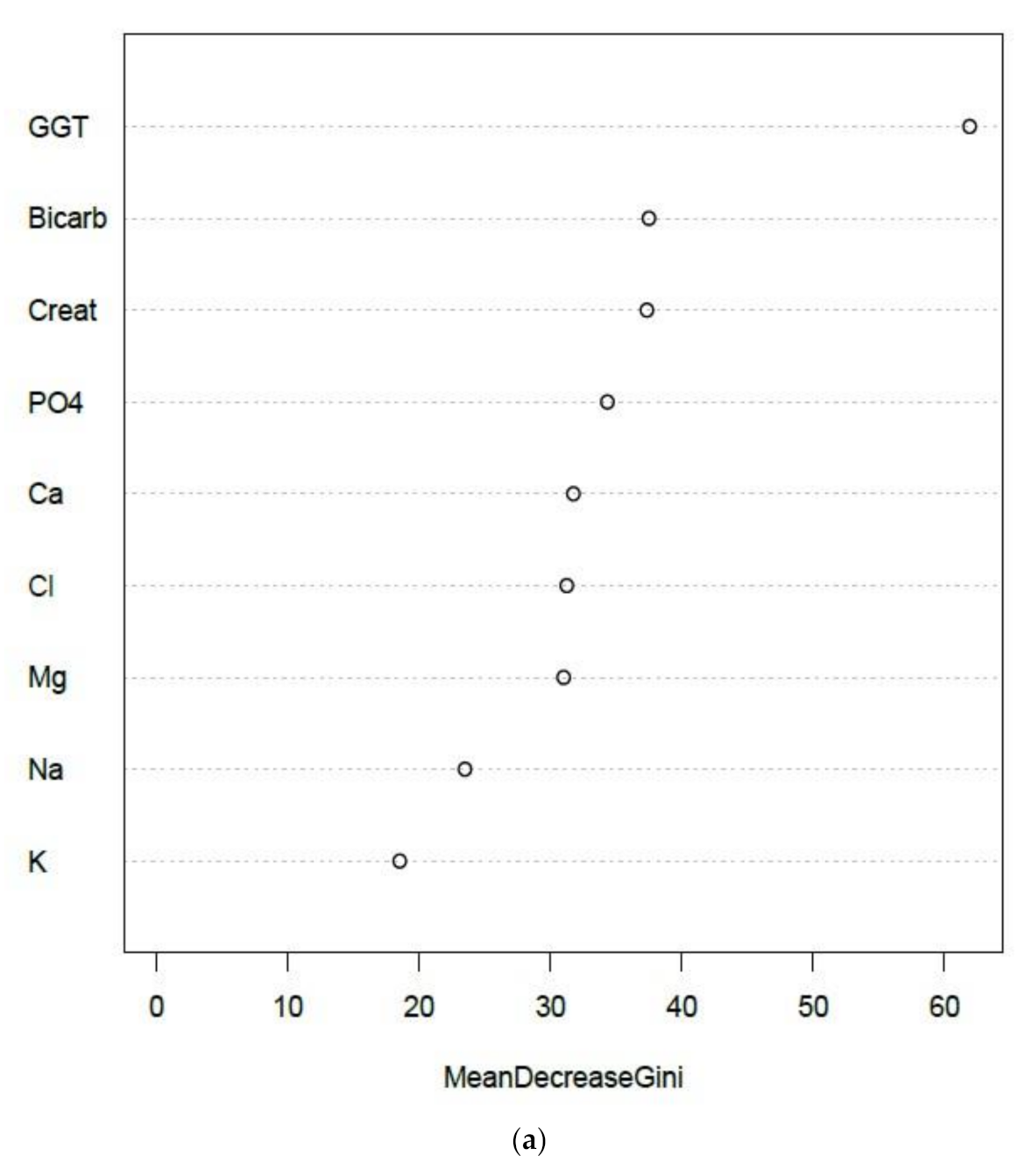
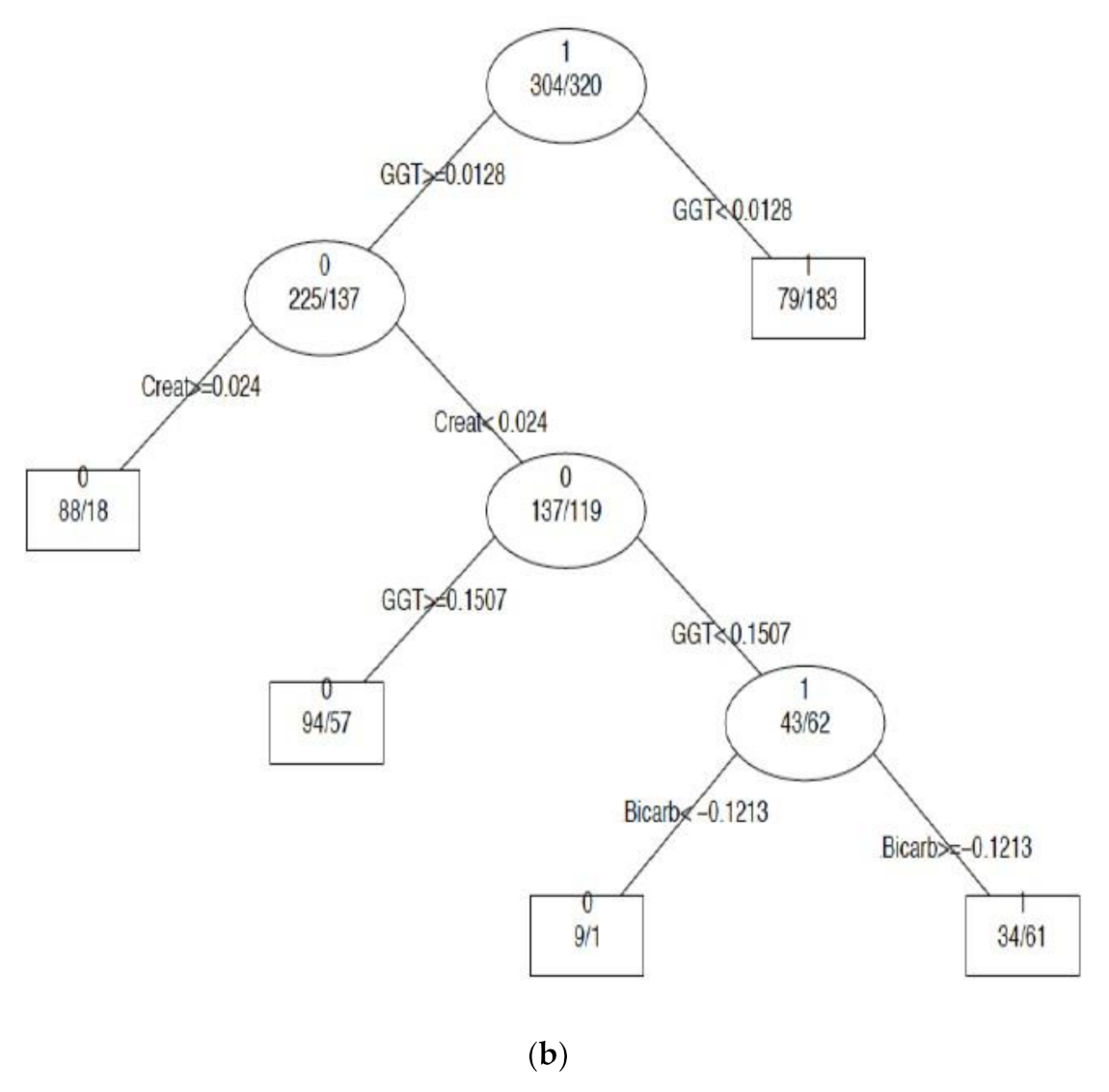

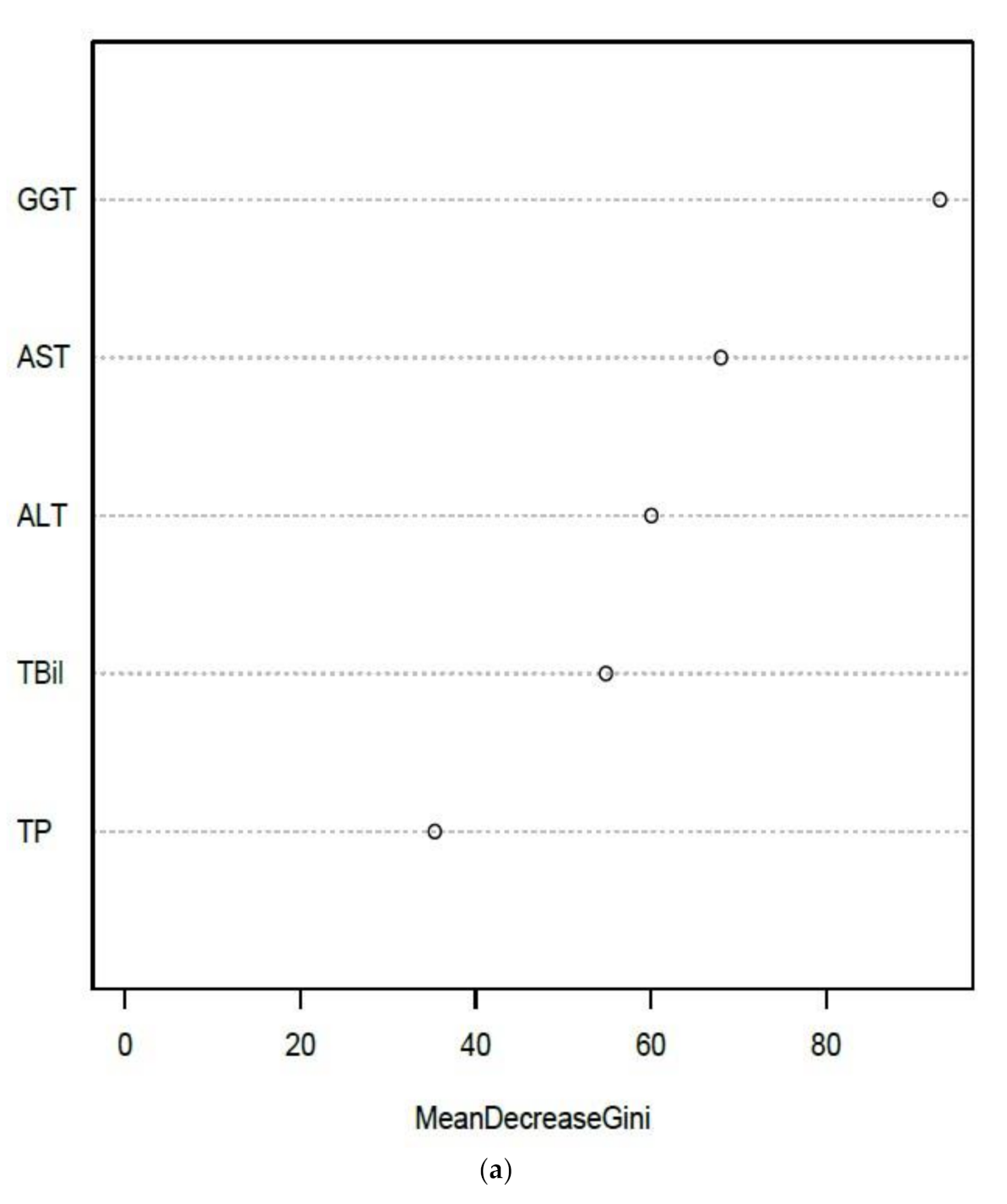

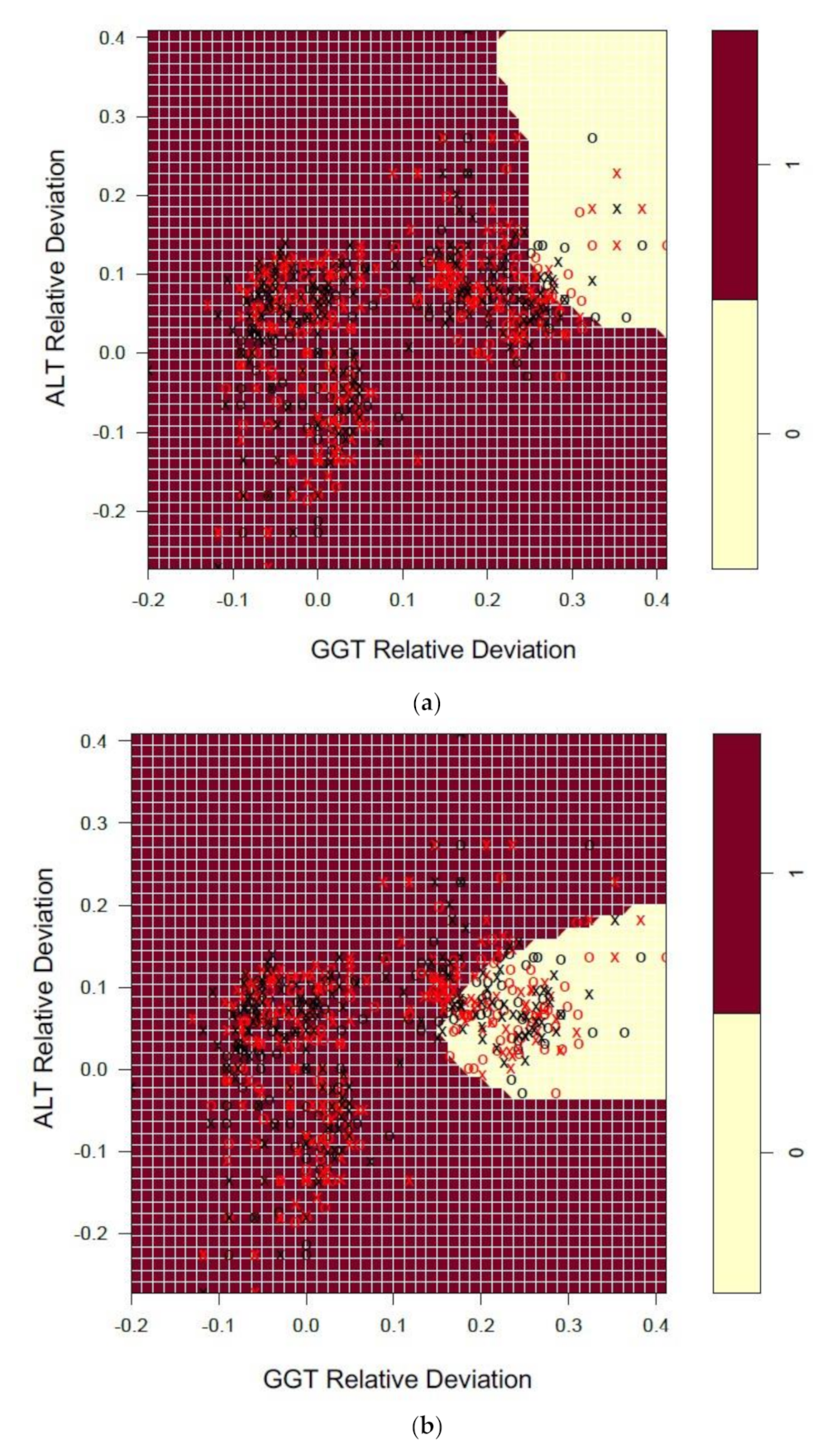
| EQA Marker | Percent (%) Quartile | NATA Conditions Quartile Means ) (± SEM) | ||
|---|---|---|---|---|
| Minor (M) | Major (C) | Minor + Major | ||
| Serum Creatinine | 1 | 3.6 ± 0.9 | 2.0 ± 1.0 | 5.6 ± 1.7 |
| 2 | 6.4 ± 0.9 | 3.8 ± 0.6 | 10.1 ± 1.4 | |
| 3 | 7.7 ± 1.4 | 4.2 ± 1.3 | 11.9 ± 2.6 | |
| 4 | 4.8 ± 1.0 | 1.0 ± 0.6 | 5.8 ± 1.3 | |
| GGT | 1 | 6.6 ± 1.0 | 5.1 ± 0.9 | 11.6 ± 1.9 |
| 2 | 6.3 ± 1.4 | 3.0 ± 1.0 | 9.3 ± 2.2 | |
| 3 | 4.0 ± 0.9 | 1.7 ± 0.5 | 5.7 ± 1.2 | |
| 4 | 7.1 ± 1.4 | 2.5 ± 1.3 | 9.6 ± 2.5 | |
| Serum Potassium | 1 | 6.3 ± 1.0 | 3.5 ± 0.9 | 9.8 ± 1.8 |
| 2 | 3.8 ± 0.9 | 1.9 ± 1.0 | 5.7 ± 1.8 | |
| 3 | 7.3 ± 1.7 | 4.3 ± 0.9 | 11.5 ± 2.3 | |
| 4 | 6.4 ± 1.2 | 3.0 ± 1.0 | 9.4 ± 2.2 | |
| Features and Results from RF Models * | Random Forest (RF) | |||
|---|---|---|---|---|
| Optimal mtry | Sensitivity | Specificity | Final Model Accuracy | |
| Model Tuning | 4 | 0.74 | 0.65 | 0.71 (95% CI: 0.64, 0.78) |
| Final Model Statistics | McNemar’s | Sensitivity | Specificity | |
| 0.27 | 0.77 | 0.65 | ||
| Kappa | PPV | NPV | ||
| 0.42 | 0.71 | 0.72 | ||
| NATA Class- Minor Reports | GGT Included in UEC Model | GGT Excluded from UEC Model | ||
|---|---|---|---|---|
| Prediction Accuracy (%) | Overall Accuracy (%) | Prediction Accuracy (%) | Overall Accuracy (%) | |
| High (>Median) | 241/320 (75.3%) | 71.3% | 230/320 (71.9%) | 66.4% |
| Low (<Median) | 198/296 (66.9%) | 179/296 (60.5%) | ||
| Features and Results from RF Models * | Random Forest (RF) | |||
|---|---|---|---|---|
| Optimal mtry | Sensitivity | Specificity | Final Model Accuracy | |
| Model Tuning | 4 | 0.75 | 0.71 | 0.72 (95% CI: 0.65, 0.78) |
| Final Model Statistics | McNemar’s | Sensitivity | Specificity | |
| 0.27 | 0.77 | 0.66 | ||
| Kappa | PPV | NPV | ||
| 0.43 | 0.71 | 0.73 | ||
| Features and Results from RF Models | Random Forest (RF) | |||
|---|---|---|---|---|
| Optimal mtry | Sensitivity | Specificity | Final Model Accuracy | |
| Model Tuning | 2 | 0.73 | 0.66 | 0.71 (95% CI: 0.64, 0.78) |
| Final Model Statistics | McNemar’s | Sensitivity | Specificity | |
| 0.055 | 0.80 | 0.61 | ||
| Kappa | PPV | NPV | ||
| 0.42 | 0.69 | 0.74 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lidbury, B.A.; Koerbin, G.; Richardson, A.M.; Badrick, T. Gamma-Glutamyl Transferase (GGT) Is the Leading External Quality Assurance Predictor of ISO15189 Compliance for Pathology Laboratories. Diagnostics 2021, 11, 692. https://doi.org/10.3390/diagnostics11040692
Lidbury BA, Koerbin G, Richardson AM, Badrick T. Gamma-Glutamyl Transferase (GGT) Is the Leading External Quality Assurance Predictor of ISO15189 Compliance for Pathology Laboratories. Diagnostics. 2021; 11(4):692. https://doi.org/10.3390/diagnostics11040692
Chicago/Turabian StyleLidbury, Brett A., Gus Koerbin, Alice M. Richardson, and Tony Badrick. 2021. "Gamma-Glutamyl Transferase (GGT) Is the Leading External Quality Assurance Predictor of ISO15189 Compliance for Pathology Laboratories" Diagnostics 11, no. 4: 692. https://doi.org/10.3390/diagnostics11040692
APA StyleLidbury, B. A., Koerbin, G., Richardson, A. M., & Badrick, T. (2021). Gamma-Glutamyl Transferase (GGT) Is the Leading External Quality Assurance Predictor of ISO15189 Compliance for Pathology Laboratories. Diagnostics, 11(4), 692. https://doi.org/10.3390/diagnostics11040692






