Abstract
Here, we develop a robust and sensitive real-time PCR assay which allows the simultaneous detection of vanA and vanB genes using common primers. The system was designed using the Primer3 online software. The specificity of primers and probes was first checked by in silico PCR and by BlastN analysis. The genomic DNA of 255 bacterial isolates, including Enterococcus spp., Gram-negative, and Gram-positive strains, as well as a collection of 50 stool and 50 rectal swab samples, were tested to evaluate the specificity of the new real-time PCR (RT-PCR) system. The results of the designed RT-PCR were 100% specific and 100% positive on tested vancomycin resistant isolates harboring either the vanA or vanB gene. RT-PCR assays were negative for all other bacterial species tested including vancomycin-sensitive Enterococci and Enterococcus strains harboring vanC genes. The limit of detection of vanA and vanB genes by RT-PCR assay was 47 CFU/mL and 32 CFU/mL, respectively. The rapid and accurate detection of vancomycin-resistant Enterococci is the cornerstone for minimizing the risk of nosocomial transmissions and outbreaks. We believe that this assay will strengthen routine diagnostics and surveillance programs.
1. Introduction
Enterococci strains display both intrinsic and acquired resistance to many antibiotic classes, such as β-lactams, aminoglycosides, fluoroquinolones, and glycopeptides [1]. In the late 1970s, the first vancomycin-resistant enterococci (VRE) strains were isolated and, since then, they have spread rapidly worldwide [2], dramatically reducing therapeutic options against Enterococci infections [3]. E. faecalis and E. faecium are the main causative agents for serious nosocomial infections in healthcare settings [4]. Resistance to vancomycin is more frequently encountered in E. faecium than E. faecalis [5]. The resistance genotypes vanA and vanB are the most frequently encountered in invasive infections and nosocomial outbreaks [6,7,8]. It is well established that the vanA gene confers a high resistance to vancomycin and teicoplanin while the vanB gene confers low-level resistance only to vancomycin, which often compromises their detection by phenotypic methods [9]. VRE infections and outbreaks are responsible for antibiotic treatment failures, an increase in morbidity and mortality, prolonged hospital stays, and high healthcare costs [10]. Active surveillance based on rapid and accurate detection of VRE isolates in high-risk patients remains the most effective strategy to reducing all these risks [3]. However, monitoring programs based on phenotypic methods or convention PCR assays are difficult to monitor optimally. However, molecular techniques based on RT-PCR effectively and robustly support these monitoring programs by avoiding the time-consuming and tedious procedures [11]. Herein, we describe for the first time a RT-PCR assay, allowing the simultaneous detection in the same reaction mixture of both vanA and vanB genes using a common set of primers and two specific probes for vanA and vanB differentiation.
2. Materials and Methods
2.1. Design of Primers and Probes
Nucleotide sequences alignment was performed using the ClustalW programme. The specificity of the selected primers and probes was firstly checked using the online “in silico PCR” programme (http://insilico.ehu.es/PCR/ accessed on 30 March 2021) and, secondly, using a BlastN analysis.
2.2. DNA Extraction
The sensitivity and specificity of our system were evaluated on the extracted genomic DNA of 255 bacterial isolates, including Enterococcus spp., Gram-negative, and Gram-positive strains. One hundred of negative stool (50) and rectal swab (50) samples were also tested. According to the manufacturer’s instructions, DNA was extracted by the EZ1 biorobot (Qiagen, Germantown, MD, USA) with the EZ1 DNA tissues kit from samples and bacterial strains. The stools and rectal swabs were firstly incubated in proteinase K solution at 56°C for four hours. The genomic DNA of bacterial strains was extracted directly from three to five colonies cultured for 24 h on Columbia agar (bioMérieux, Marcy l’Étoile, France).
2.3. RT-PCR Assay
The sensitivity of our RT-PCR assay was determined using a series of 10-fold dilutions from an initial inoculum at 106 colony-forming units/mL (CFU/mL) of both E. faecium (DSMZ 17050) and E. faecalis (DSMZ 12956) reference strains, as positive controls, carrying the vanA and vanB genes, respectively. The efficiency parameters, slope, and R2 were calculated using a standard curve efficiency established on the basis of the obtained number of log10 CFU/mL and cycle threshold (Ct) values. The limit of detection (LoD) was based on the last dilution detected by an RT-PCR reaction before 35 cycles. To perform the RT-PCR reaction, we used a CFX96 device connected with TM-BioRad using TaqMan technology (Bio-Rad, Lab. Inc., Hercules, CA, USA). The RT-PCR assay conditions were as follows: 50 °C for 2 min, 95 °C for 15 min, and 35 cycles of 95 °C for 1 s and 62 °C for 30 s.
The assay was also tested in an ex-vivo spiking experiment to evaluate their sensitivity and specificity in which negative stool and rectal swab samples were spiked with reference strains carrying vanA and vanB genes. Five stool samples and five rectal swabs were dissolved in 10 mL of sterile brain heart infusion broth (BHI) (Laboratoire Conda S.A, Torrejón de Ardoz, Spain). Each sample was inoculated using a series of 10-fold dilutions from an initial inoculum at 106 of E. faecium (DSMZ 17050) and E. faecalis (DSMZ 12956). The DNA extraction and RT-PCR assays were performed from different concentrations, as described above.
3. Results
3.1. Design of Primers and Probes
Nucleotide sequences of vanA and vanB variant genes were collected from the GenBank database and aligned. As shown in Figure 1, identified sequences discriminated between both genes using different internal bases, which were used to pick the specific probe for each gene. A conserved sequence of 160-bp between vanA and vanB genes was identified, then conserved, and common bases of both ends were then used as the annealing sites of common primers (Figure 1). The designed primers and probes were checked in silico to avoid misleading amplification of vanC gene variants.

Figure 1.
Primer and probe sequences alignment and selected sequence targets. (A) sequence alignment of van genes for selection of primers and probes; (B) Table presenting the selected primers and probes with the size (bp) of each sequence and PCR product.
3.2. Specificity and Sensitivity Tests of the Designed RT-PCR Assay
From extracted genomic DNA of 255 bacterial strains, including 191 Enterococcus spp., 26 Gram-negative, 38 Gram-positive strains, and 100 stool and rectal swab specimens, the RT-PCR results were 100% specific and 100% positive on tested vancomycin resistant isolates harboring either vanA or vanB gene (Table 1). No false-positive was detected by the RT-PCR assay.

Table 1.
List of the clinical isolates tested for the specificity of the designed RT-PCR assay.
This RT-PCR was negative for all tested samples and other bacterial species tested, including vancomycin-sensitive Enterococci strains harboring the vanC gene. The detection limit of the vanA gene in E. faecium (DSMZ 17050) and the vanB gene in E. faecalis (DSMZ 12956) was 47 CFU/mL and 32 CFU/mL, respectively. Finally, the results showed the same limit of detection (LOD) value, for both vanA and vanB genes in samples spiked ex-vivo even in the presence of DNA from other microbes, which are naturally present in stools and rectums. As shown in Figure 2, the dynamic range of the amplification reaction of genomic DNA dilutions of both control strains spanned up to 101 CFU/mL, displaying a correlation coefficient (R²) of 0.9919 for vanA and 0.9916 for vanB genes. The amplification curves were linear and strongly correlated with corresponding Ct values. Replication efficiency was 98.38% and 132.23% with a slope of −3.3614 and −2.7329 for the vanA and vanB genes, respectively.
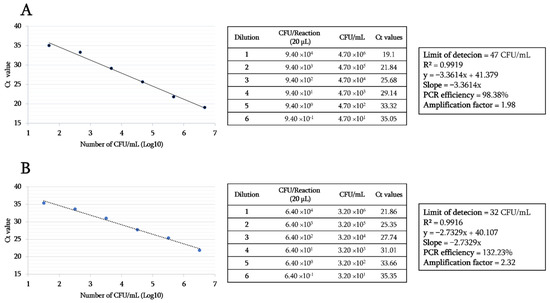
Figure 2.
Sensitivity test and limit of detection of vanA-vanB real-time PCR assay. (A) the amplification curves and detection details of the vanA gene. (B) the amplification curves and detection details of the vanB gene.
4. Discussion
Within hospitals, VRE spreads rapidly through patients, caregivers and the environment [12]. Horizontal gene transfer of the van operon between Enterococcus spp. strains and/or other bacteria can occur frequently. For these reasons, the presence of VREs must be rapidly controlled in health care facilities [4]. Molecular methods have shown to have several advantages compared to phenotypic methods for the detection and distinction of vancomycin-resistance genotypes [11,13]. Moreover, our RT-PCR assay presents a new method that could simultaneously amplify both vanA and vanB genes in the same mixture reaction. Clostridium spp. commonly found in the human gastrointestinal tract that may harbor the different vanB variants [14] can induce false-positive reactions in commercial diagnostic kits. It is essential to highlight that, in this study, we tested a large collection of strains, including several bacterial species, including Clostridium spp. and also clinical samples (n = 100), especially stools and rectal swabs, making the percentage of specificity much more relevant. This system represents a considerable economy in comparison with other RT-PCR systems described previously [9,11,15,16], which required specific primers pair for each gene. The reduced costs and time for diagnostic results are crucial in the management of nosocomial outbreaks. The other molecular support of glycopeptides resistance conferring a low level of resistance to vancomycin, such as the vanC gene, is not detected by this system, which could be considered a limitation of this method.
5. Conclusions
We believe that our system may be beneficial for hospital settings receiving many critically ill patients and microbiology laboratories in the detection of Enterococcus isolates bearing either the vanA or vanB gene.
Author Contributions
Conceptualization S.-A.R., S.M.D. and J.-M.R. methodology: H.Z, S.M.D. formal analysis: H.Z., L.H, and S.M.D. writing—original draft preparation: H.Z. writing—review & editing: H.Z., S.-A.R., L.H., J.-M.R., and S.M.D. supervision: J.-M.R.; and S.M.D. funding acquisition: J.-M.R.; and S.M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Algerian Ministry of Higher Education and Scientific Research and by the French Government under the “Investissements d’avenir” (reference: Méditerranée Infection 10-IAHU-03).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are very grateful to the staff of Tlemcen University Hospital for helping with sample collection and for providing information.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Faron, M.L.; Ledeboer, N.A.; Buchan, B.W. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J. Clin. Microbiol. 2016, 54, 2436–2447. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, H. Controlling the spread of vancomycin-resistant enterococci. Is active screening worthwhile? J. Hosp. Infect. 2014, 88, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Reyes, K.; Bardossy, A.C.; Zervos, M. Vancomycin-Resistant Enterococci: Epidemiology, Infection Prevention, and Control. Infect. Dis. Clin. N. Am. 2016, 30, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Kos, V.N.; Gilmore, M.S. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 2010, 13, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Guzman Prieto, A.M.; van Schaik, W.; Rogers, M.R.C.; Coque, T.M.; Baquero, F.; Corander, J.; Willems, R.J.L. Global emergence and dissemination of enterococci as nosocomial pathogens: Attack of the clones? Front. Microbiol. 2016, 7, 788. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Tedim, A.P.; Francia, M.V.; Jensen, L.B.; Novais, C.; Peixe, L.; Sánchez-Valenzuela, A.; Sundsfjord, A.; Hegstad, K.; Werner, G.; et al. Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986–2012). J. Antimicrob. Chemother. 2016, 71, 3351–3366. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.; Ballard, S.; Sullivan, S.; Marshall, C. An outbreak of vanA vancomycin-resistant Enterococcus faecium in a hospital with endemic vanB VRE. Infect. Dis. Health 2019, 24, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Rangberg, A.; Larsen, A.L.; Kacelnik, O.; Sæther, H.S.; Bjørland, M.; Ringstad, J.; Jonassen, C.M. Molecular analysis and epidemiological typing of Vancomycin-resistant Enterococcus outbreak strains. Sci. Rep. 2019. [Google Scholar] [CrossRef] [PubMed]
- He, Y.H.; Ruan, G.J.; Hao, H.; Xue, F.; Ma, Y.K.; Zhu, S.N.; Zheng, B. Real-time PCR for the rapid detection of vanA, vanB and vanM genes. J. Microbiol. Immunol. Infect. 2019, 9, 11917. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.F.; Soumakis, S.A.; Rendo, A.; Herrington, J.A.; Gianarkis, D.G.; Thurberg, B.E.; Painter, B.G. Epidemiologic analysis and genotypic characterization of a nosocomial outbreak of vancomycin-resistant enterococci. J. Clin. Microbiol. 1993, 31, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Palladino, S.; Kay, I.D.; Flexman, J.P.; Boehm, I.; Costa, A.M.G.; Lambert, E.J.; Christiansen, K.J. Rapid detection of vanA and vanB genes directly from clinical specimens and enrichment broths by real-time multiplex PCR assay. J. Clin. Microbiol. 2003, 41, 2483–2486. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frakking, F.N.J.; Bril, W.S.; Sinnige, J.C.; van’t Klooster, J.E.; de Jong, B.A.W.; van Hannen, E.J.; Tersmette, M. Recommendations for the successful control of a large outbreak of vancomycin-resistant Enterococcus faecium in a non-endemic hospital setting. J. Hosp. Infect. 2018, 100, e216–e225. [Google Scholar] [CrossRef] [PubMed]
- Sloan, L.M.; Uhl, J.R.; Vetter, E.A.; Schleck, C.D.; Harmsen, W.S.; Manahan, J.; Thompson, R.L.; Rosenblatt, J.E.; Cockerill, F.R. Comparison of the Roche LightCycler vanA/vanB detection assay and culture for detection of vancomycin-resistant enterococci from perianal swabs. J. Clin. Microbiol. 2004, 42, 2636–2643. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Domingo, M.C.; Huletsky, A.; Bernal, A.; Giroux, R.; Boudreau, D.K.; Picard, F.J.; Bergeron, M.G. Characterization of a Tn5382-like transposon containing the vanB2 gene cluster in a Clostridium strain isolated from human faeces. J. Antimicrob. Chemother. 2005, 55, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Cekin, Y.; Daloǧlu, A.E.; Öǧünç, D.; Baysan, B.Ö.; Daǧlar, D.; Inan, D.; Mutlu, D.; Öngüt, G.; Çolak, D. Evaluation of vancomycin resistance 3 multiplexed pcr assay for detection of vancomycin-resistant enterococci from rectal swabs. Ann. Lab. Med. 2013, 33, 326–330. [Google Scholar] [CrossRef]
- Shanmugakani, R.K.; Fujiya, Y.; Akeda, Y.; Hamaguchi, S.; Hamada, S.; Tomono, K. Rapid multiplex detection of the resistance genes mecA, vanA and vanB from Gram-positive cocci-positive blood cultures using a PCR-dipstick technique. J. Med. Microbiol. 2020, 69, 249–255. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).