Deep Learning Models for Poorly Differentiated Colorectal Adenocarcinoma Classification in Whole Slide Images Using Transfer Learning
Abstract
:1. Introduction
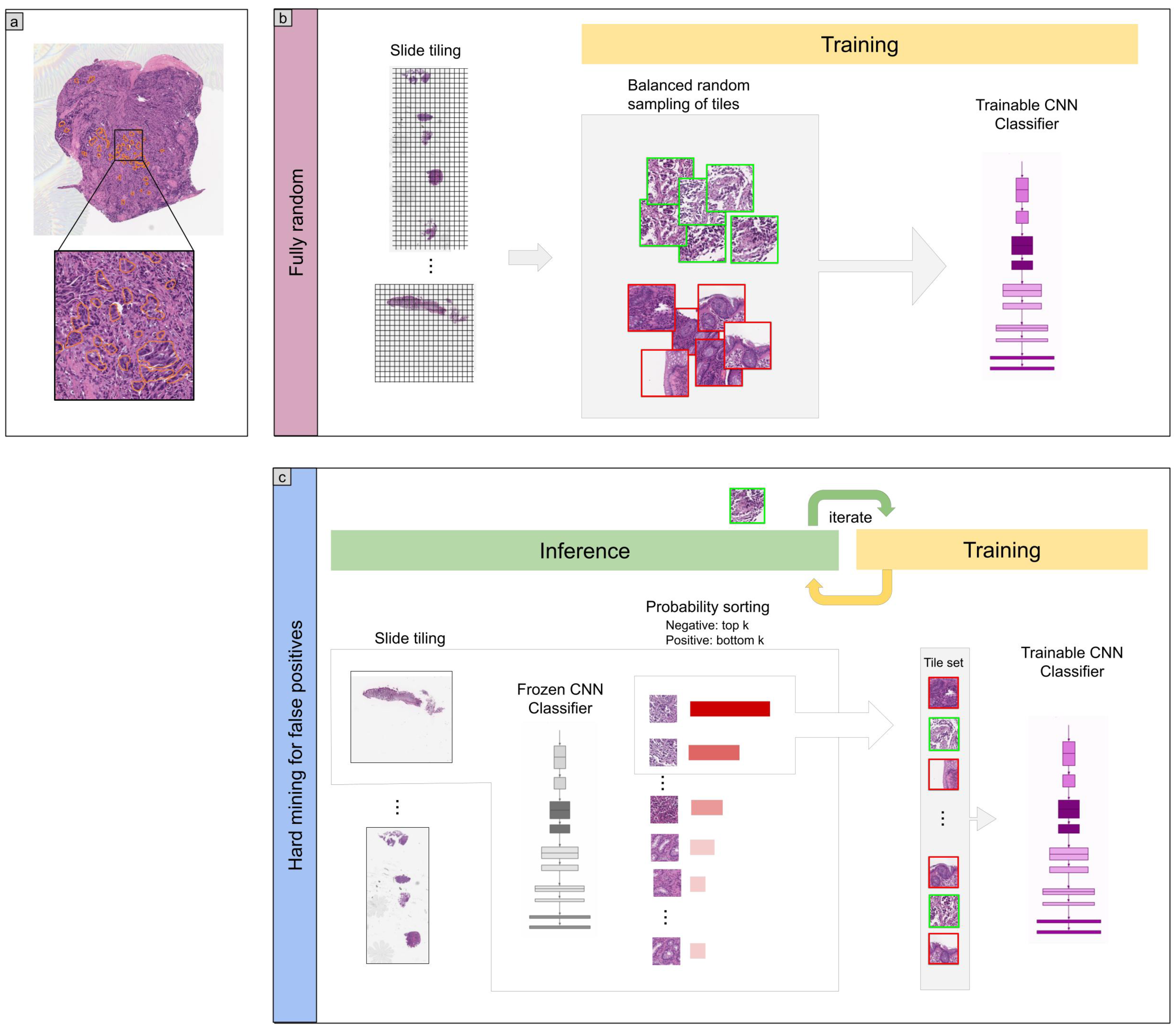
2. Methods
2.1. Clinical Cases and Pathological Records
2.2. Dataset and Annotations
2.3. Deep Learning Models
2.4. Software and Statistical Analysis
3. Results
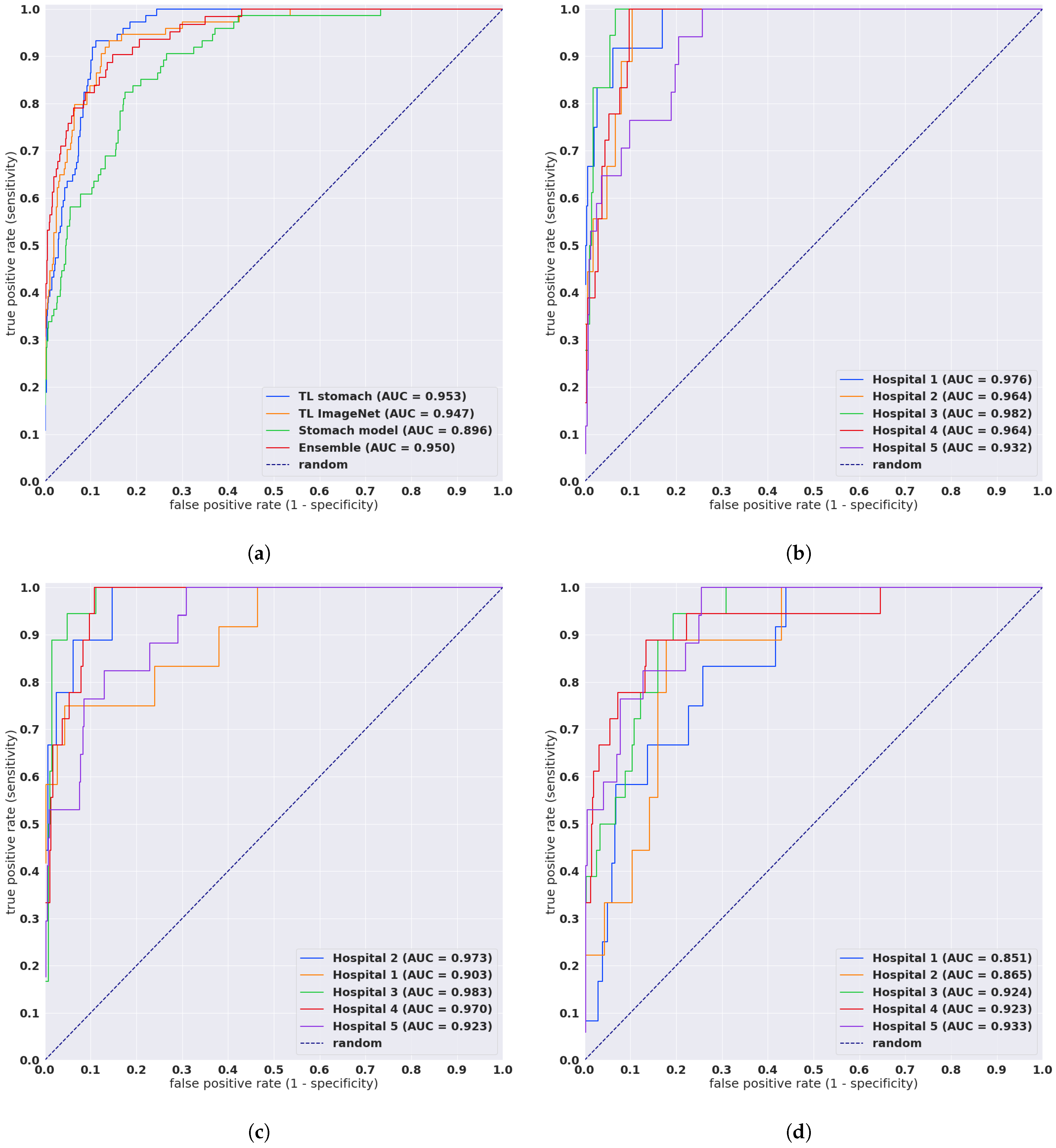
Evaluation on Five Independent Test Sets from Different Sources
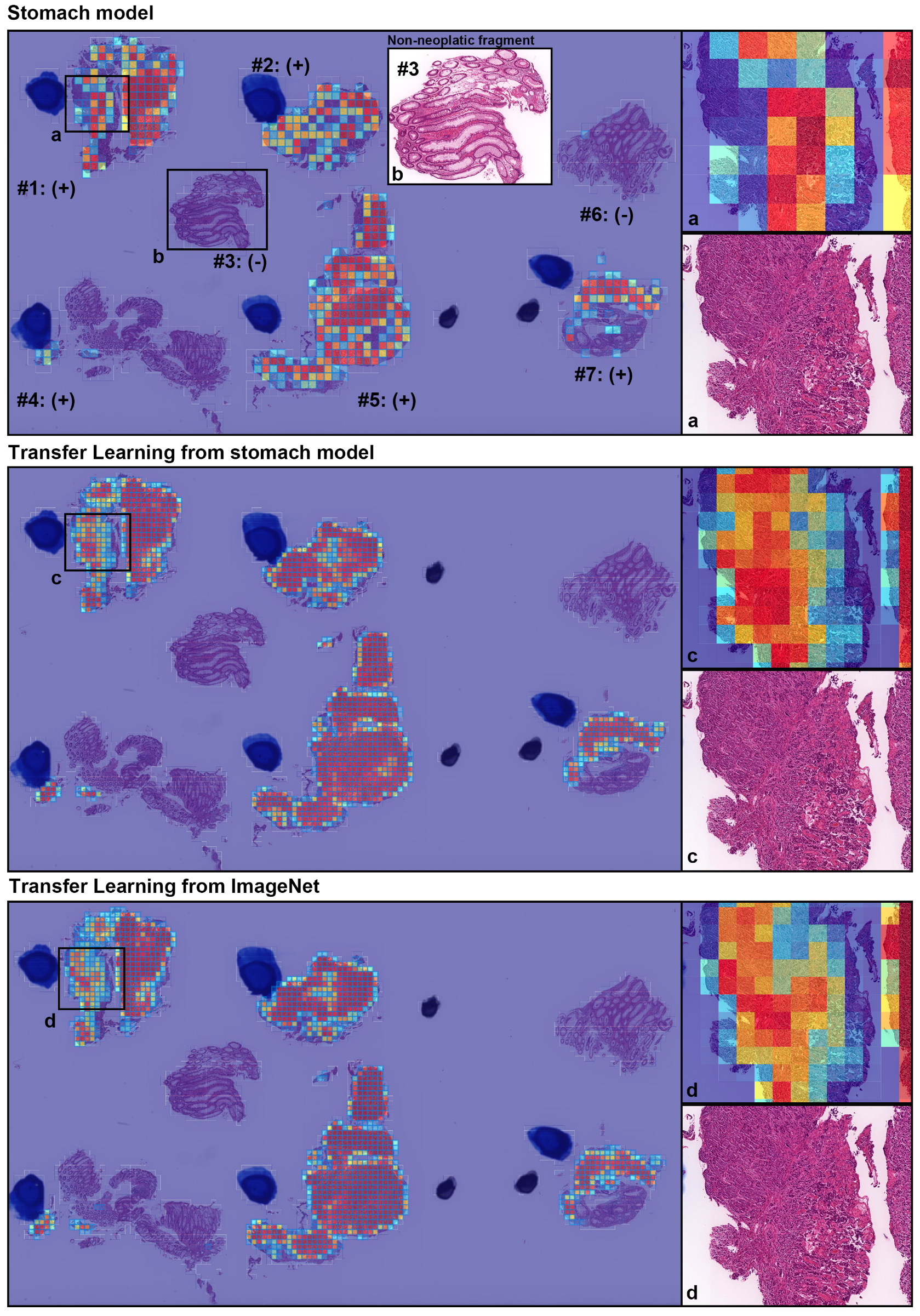
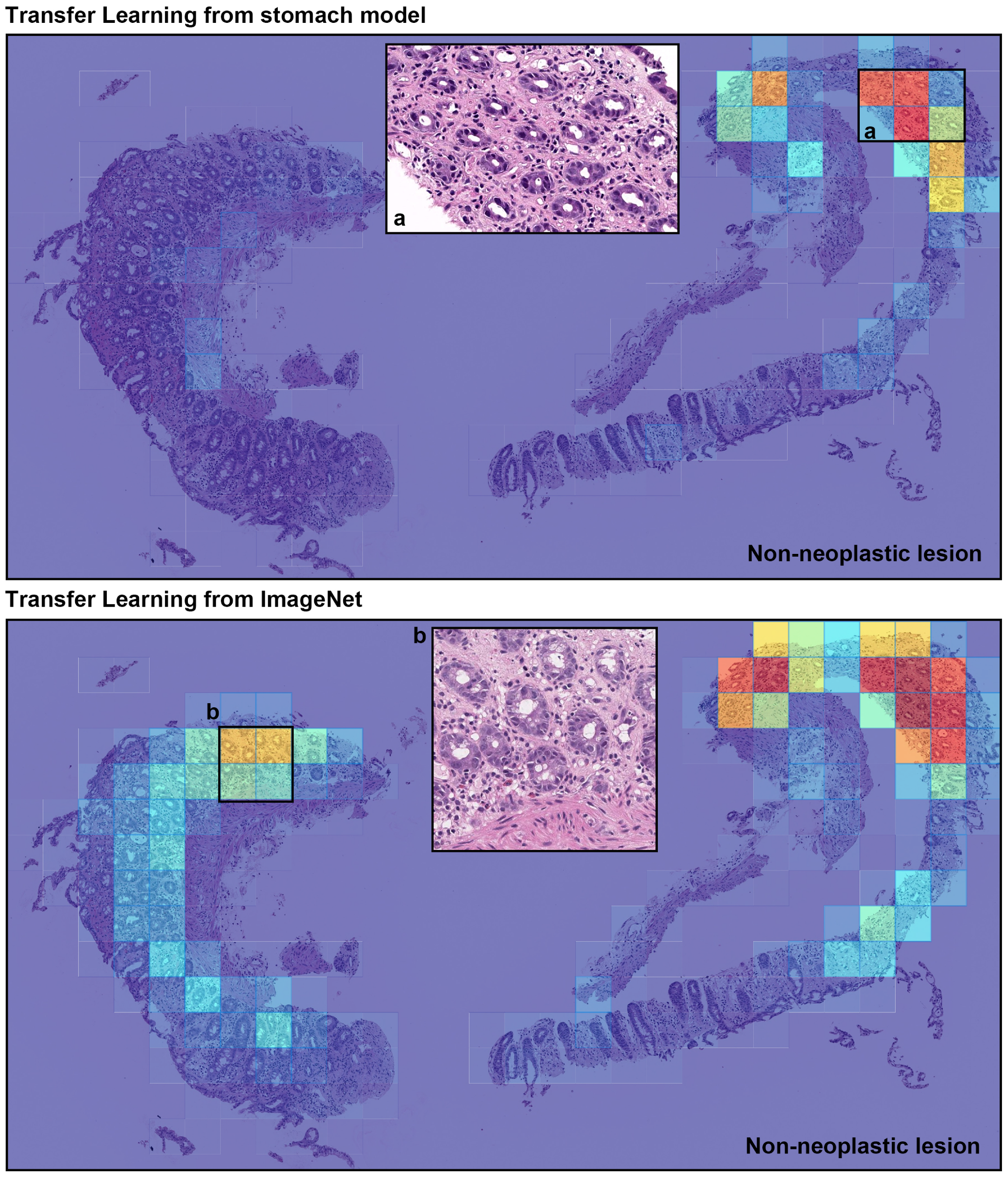
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wolf, A.M.; Fontham, E.T.; Church, T.R.; Flowers, C.R.; Guerra, C.E.; LaMonte, S.J.; Etzioni, R.; McKenna, M.T.; Oeffinger, K.C.; Shih, Y.C.T.; et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J. Clin. 2018, 68, 250–281. [Google Scholar] [CrossRef]
- Winawer, S.J.; Zauber, A.G. The advanced adenoma as the primary target of screening. Gastrointest. Endosc. Clin. N. Am. 2002, 12, 1–9. [Google Scholar] [CrossRef]
- Yu, K.H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Ré, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef] [Green Version]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-based convolutional neural network for whole slide tissue image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2424–2433. [Google Scholar]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [Green Version]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van De Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraus, O.Z.; Ba, J.L.; Frey, B.J. Classifying and segmenting microscopy images with deep multiple instance learning. Bioinformatics 2016, 32, i52–i59. [Google Scholar] [CrossRef] [PubMed]
- Korbar, B.; Olofson, A.M.; Miraflor, A.P.; Nicka, C.M.; Suriawinata, M.A.; Torresani, L.; Suriawinata, A.A.; Hassanpour, S. Deep learning for classification of colorectal polyps on whole-slide images. J. Pathol. Inform. 2017, 8, 30. [Google Scholar] [PubMed]
- Luo, X.; Zang, X.; Yang, L.; Huang, J.; Liang, F.; Rodriguez-Canales, J.; Wistuba, I.I.; Gazdar, A.; Xie, Y.; Xiao, G. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J. Thorac. Oncol. 2017, 12, 501–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Tafe, L.J.; Linnik, Y.A.; Vaickus, L.J.; Tomita, N.; Hassanpour, S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 2019, 9, 3358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gertych, A.; Swiderska-Chadaj, Z.; Ma, Z.; Ing, N.; Markiewicz, T.; Cierniak, S.; Salemi, H.; Guzman, S.; Walts, A.E.; Knudsen, B.S. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 2019, 9, 1483. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018, 23, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 2020, 10, 1504. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, S.; Vogelstein, B.; Kudo, S.; Riboli, E.; Nakamura, S.; Hainaut, P. Carcinoma of the colon and rectum. Pathology and genetics of tumours of the digestive system. In World Health Organization Classification of Tumours; IARC Press: Lyon, France, 2000; pp. 103–119. [Google Scholar]
- Ogawa, M.; Watanabe, M.; Eto, K.; Kosuge, M.; Yamagata, T.; Kobayashi, T.; Yamazaki, K.; Anazawa, S.; Yanaga, K. Poorly differentiated adenocarcinoma of the colon and rectum: Clinical characteristics. Hepato-gastroenterology 2008, 55, 907–911. [Google Scholar]
- Winn, B.; Tavares, R.; Matoso, A.; Noble, L.; Fanion, J.; Waldman, S.A.; Resnick, M.B. Expression of the intestinal biomarkers Guanylyl cyclase C and CDX2 in poorly differentiated colorectal carcinomas. Hum. Pathol. 2010, 41, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Kazama, Y.; Watanabe, T.; Kanazawa, T.; Tanaka, J.; Tanaka, T.; Nagawa, H. Poorly differentiated colorectal adenocarcinomas show higher rates of microsatellite instability and promoter methylation of p16 and hMLH1: A study matched for T classification and tumor location. J. Surg. Oncol. 2008, 97, 278–283. [Google Scholar] [CrossRef]
- Takeuchi, K.; Kuwano, H.; Tsuzuki, Y.; Ando, T.; Sekihara, M.; Hara, T.; Asao, T. Clinicopathological characteristics of poorly differentiated adenocarcinoma of the colon and rectum. Hepato-gastroenterology 2004, 51, 1698–1702. [Google Scholar] [PubMed]
- Secco, G.; Fardelli, R.; Campora, E.; Lapertosa, G.; Gentile, R.; Zoli, S.; Prior, C. Primary mucinous adenocarcinomas and signet-ring cell carcinomas of colon and rectum. Oncology 1994, 51, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Taniyama, K.; Suzuki, H.; Matsumoto, M.; Hakamada, K.; Toyam, K.; Tahara, E. Flow Cytometric DNA Analysis of Poorly Differentialted Adenocarcinoma of the Colorectum. Jpn. J. Clin. Oncol. 1991, 21, 406–411. [Google Scholar] [PubMed]
- Kawabata, Y.; Tomita, N.; Monden, T.; Ohue, M.; Ohnishi, T.; Sasaki, M.; Sekimoto, M.; Sakita, I.; Tamaki, Y.; Takahashi, J.; et al. Molecular characteristics of poorly differentiated adenocarcinoma and signet-ring-cell carcinoma of colorectum. Int. J. Cancer 1999, 84, 33–38. [Google Scholar] [CrossRef]
- Sugao, Y.; Yao, T.; Kubo, C.; Tsuneyoshi, M. Improved prognosis of solid-type poorly differentiated colorectal adenocarcinoma: A clinicopathological and immunohistochemical study. Histopathology 1997, 31, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Komori, K.; Kanemitsu, Y.; Ishiguro, S.; Shimizu, Y.; Sano, T.; Ito, S.; Abe, T.; Senda, Y.; Misawa, K.; Ito, Y.; et al. Clinicopathological study of poorly differentiated colorectal adenocarcinomas: Comparison between solid-type and non-solid-type adenocarcinomas. Anticancer Res. 2011, 31, 3463–3467. [Google Scholar] [PubMed]
- Kanavati, F.; Tsuneki, M. Partial transfusion: On the expressive influence of trainable batch norm parameters for transfer learning. arXiv 2021, arXiv:2102.05543. [Google Scholar]
- Kanavati, F.; Tsuneki, M. A deep learning model for gastric diffuse-type adenocarcinoma classification in whole slide images. arXiv 2021, arXiv:2104.12478. [Google Scholar]
- Kanavati, F.; Toyokawa, G.; Momosaki, S.; Rambeau, M.; Kozuma, Y.; Shoji, F.; Yamazaki, K.; Takeo, S.; Iizuka, O.; Tsuneki, M. Weakly-supervised learning for lung carcinoma classification using deep learning. Sci. Rep. 2020, 10, 9297. [Google Scholar] [CrossRef]
- Kanavati, F.; Toyokawa, G.; Momosaki, S.; Takeoka, H.; Okamoto, M.; Yamazaki, K.; Takeo, S.; Iizuka, O.; Tsuneki, M. A deep learning model for the classification of indeterminate lung carcinoma in biopsy whole slide images. Sci. Rep. 2021, 11, 8110. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: tensorflow.org (accessed on 24 January 2020).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T.; the scikit-image contributors. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; CRC Press: Boca Raton, FL, USA, 1994. [Google Scholar]
- Goode, A.; Gilbert, B.; Harkes, J.; Jukic, D.; Satyanarayanan, M. OpenSlide: A vendor-neutral software foundation for digital pathology. J. Pathol. Inform. 2013, 4, 27. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Zhang, C.; Ma, Y. Ensemble Machine Learning: Methods and Applications; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
| Poorly Diff. ADC | Well-to-Moderately-Diff. ADC | Adenoma | Non-Neoplastic | Total | ||
|---|---|---|---|---|---|---|
| Test | Hospital 1 | 12 | 125 | 61 | 251 | 449 |
| Hospital 2 | 9 | 41 | 78 | 44 | 172 | |
| Hospital 3 | 18 | 20 | 210 | 39 | 287 | |
| Hospital 4 | 18 | 74 | 239 | 158 | 489 | |
| Hospital 5 | 17 | 144 | 55 | 186 | 402 | |
| Training | Hospital 1 & 5 | 30 | 60 | 90 | 500 | 680 |
| Validation | Hospital 1 & 5 | 6 | 11 | 20 | 31 | 68 |
| Method | Source | ROC AUC | Log Loss |
|---|---|---|---|
| Ensemble | combined | 0.950 [0.925, 0.971] | 0.135 [0.122, 0.148] |
| TL stomach | combined | 0.953 [0.937, 0.966] | 0.466 [0.426, 0.506] |
| TL ImageNet | combined | 0.947 [0.923, 0.968] | 0.555 [0.511, 0.594] |
| Stomach model | combined | 0.896 [0.862, 0.923] | 0.863 [0.814, 0.911] |
| TL stomach | Hospital 1 | 0.976 [0.936, 0.997] | 0.236 [0.196, 0.271] |
| TL stomach | Hospital 2 | 0.964 [0.927, 0.991] | 0.459 [0.347, 0.576] |
| TL stomach | Hospital 3 | 0.982 [0.966, 0.995] | 0.195 [0.143, 0.244] |
| TL stomach | Hospital 4 | 0.964 [0.94, 0.983] | 0.44 [0.36, 0.515] |
| TL stomach | Hospital 5 | 0.932 [0.886, 0.97] | 0.949 [0.855, 1.081] |
| TL ImageNet | Hospital 1 | 0.903 [0.774, 0.993] | 0.325 [0.284, 0.367] |
| TL ImageNet | Hospital 2 | 0.973 [0.939, 0.999] | 0.613 [0.468, 0.72] |
| TL ImageNet | Hospital 3 | 0.983 [0.965, 0.997] | 0.268 [0.209, 0.326] |
| TL ImageNet | Hospital 4 | 0.97 [0.948, 0.987] | 0.48 [0.398, 0.549] |
| TL ImageNet | Hospital 5 | 0.923 [0.868, 0.969] | 1.085 [0.972, 1.219] |
| Stomach model | Hospital 1 | 0.851 [0.739, 0.928] | 1.055 [0.953, 1.167] |
| Stomach model | Hospital 2 | 0.865 [0.768, 0.951] | 0.882 [0.722, 1.032] |
| Stomach model | Hospital 3 | 0.924 [0.864, 0.96] | 0.607 [0.506, 0.716] |
| Stomach model | Hospital 4 | 0.923 [0.843, 0.981] | 0.554 [0.475, 0.62] |
| Stomach model | Hospital 5 | 0.933 [0.881, 0.972] | 1.2 [1.102, 1.326] |
| Predicted Label | |||
|---|---|---|---|
| Other | Poorly ADC | ||
| True label | Other | 1572 | 153 |
| Poorly ADC | 11 | 63 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuneki, M.; Kanavati, F. Deep Learning Models for Poorly Differentiated Colorectal Adenocarcinoma Classification in Whole Slide Images Using Transfer Learning. Diagnostics 2021, 11, 2074. https://doi.org/10.3390/diagnostics11112074
Tsuneki M, Kanavati F. Deep Learning Models for Poorly Differentiated Colorectal Adenocarcinoma Classification in Whole Slide Images Using Transfer Learning. Diagnostics. 2021; 11(11):2074. https://doi.org/10.3390/diagnostics11112074
Chicago/Turabian StyleTsuneki, Masayuki, and Fahdi Kanavati. 2021. "Deep Learning Models for Poorly Differentiated Colorectal Adenocarcinoma Classification in Whole Slide Images Using Transfer Learning" Diagnostics 11, no. 11: 2074. https://doi.org/10.3390/diagnostics11112074
APA StyleTsuneki, M., & Kanavati, F. (2021). Deep Learning Models for Poorly Differentiated Colorectal Adenocarcinoma Classification in Whole Slide Images Using Transfer Learning. Diagnostics, 11(11), 2074. https://doi.org/10.3390/diagnostics11112074







