Ex Vivo Vibration Spectroscopic Analysis of Colorectal Polyps for the Early Diagnosis of Colorectal Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Colorectal Tissue Samples
2.2. Spectroscopic Measurements
2.3. Statistical Analyses of Spectroscopic Data
3. Results
3.1. Vibrational Spectra of Normal Colon Tissues, and Adenomatous and Adenocarcinomatous Polyps
3.1.1. FTIR Spectra (900–3750 cm−1)
3.1.2. FT Raman Spectra (500–3100 cm−1)
3.1.3. Dispersion Raman Spectra (450–3100 cm−1)
3.2. Discrimination of Normal Colon Tissues, and Adenomatous and Adenocarcinomatous Polyps
3.2.1. PCA/FTIR (900–1750 cm−1 and 2830–3100 cm−1)
3.2.2. PCA/FT Raman (1190–1750 cm−1 and 2820–3100 cm−1)
3.2.3. PCA/Dispersion Raman (950–1750 cm−1 and 2750–3100 cm−1)
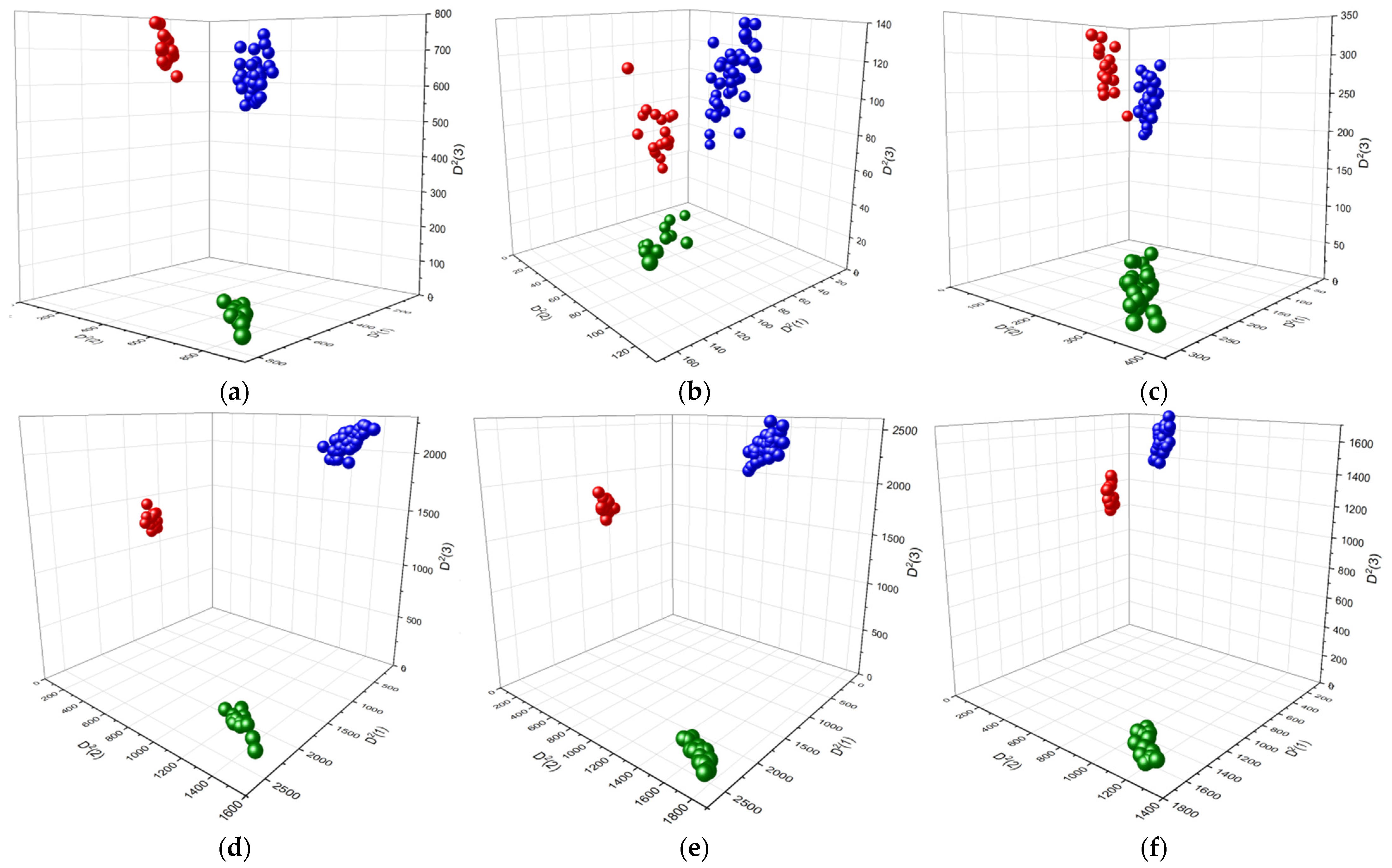
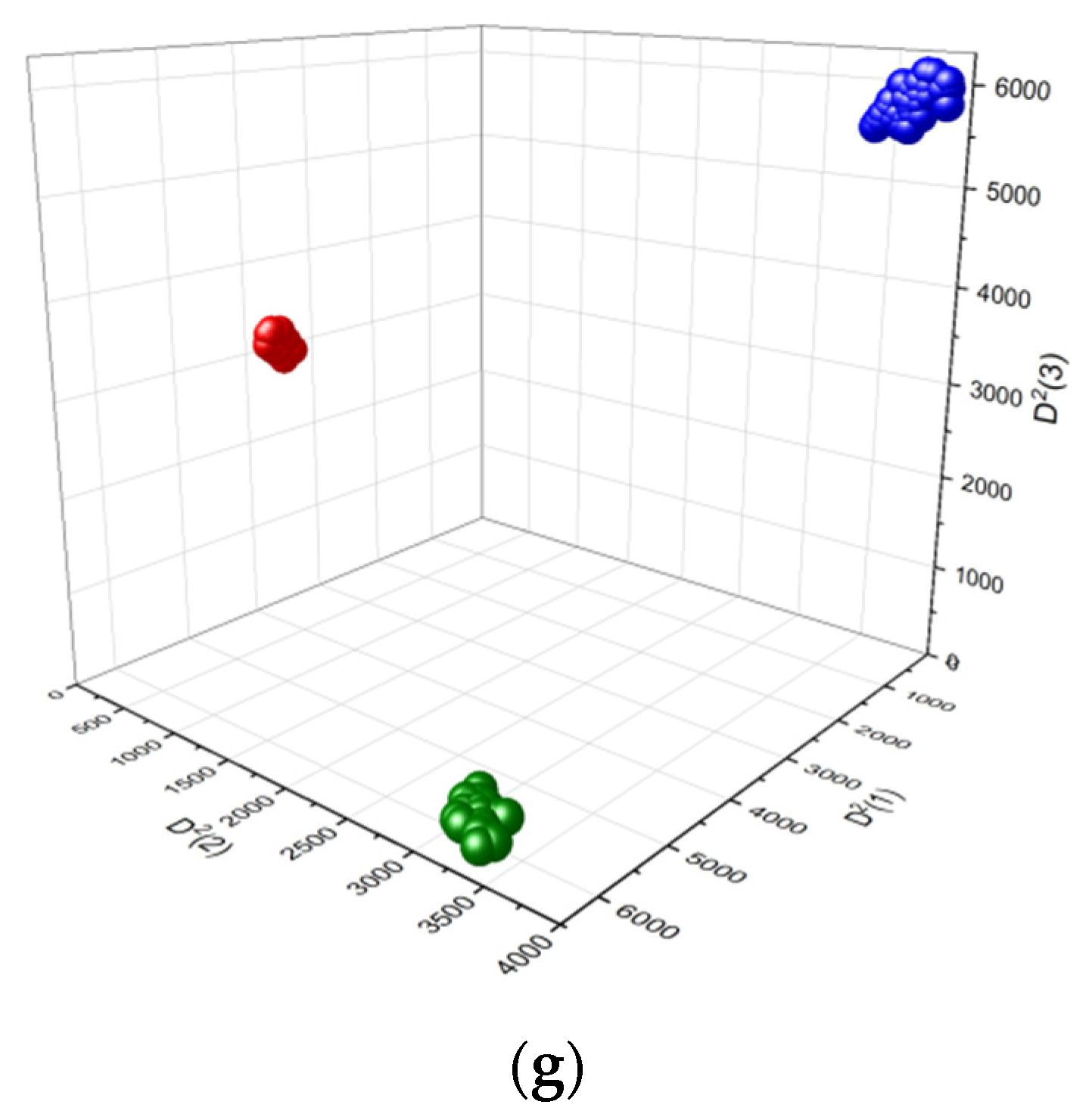
3.2.4. LDA Classification of Colon Tissues/Polyps Using Vibrational Spectroscopic Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Ciocalteu, A.; Gheonea, D.I.; Saftoiu, A.; Streba, L.; Dragoescu, N.A.; Tenea-Cojan, T.S. Current strategies for malignant pedunculated colorectal polyps. World J. Gastrointest. Oncol. 2018, 10, 465. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, M.S.; Lynch, G.; Park, J.; McSorley, S.; Edwards, J. Novel methods of risk stratifying patients for metachronous, pre-malignant colorectal polyps: A systematic review. Crit. Rev. Oncol. Hematol. 2021, 164, 103421. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of colorectal carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Aceto, G.M.; Catalano, T.; Curia, M.C. Molecular aspects of colorectal adenomas: The interplay among microenvironment, oxidative stress, and predisposition. Bio. Med. Res. Int. 2020, 2020, 1726309. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, A.; Shyne, M.; Mandel, J.S.; Snover, D.; Church, T.R. Colonoscopy with polypectomy reduces long-term incidence of colorectal cancer in both men and women: Extended results from the minnesota colon cancer control study. Gastroenterology 2021, 160, 1397–1399. [Google Scholar] [CrossRef]
- Romiti, A.; Roberto, M.; Marchetti, P.; Di Cerbo, A.; Falcone, R.; Campisi, G.; Ferri, M.; Balducci, G.; Ramacciato, G.; Ruco, L.; et al. Study of histopathologic parameters to define the prognosis of stage II colon cancer. Int. J. Colorectal Dis. 2019, 34, 905–913. [Google Scholar] [CrossRef]
- Kallenbach-Thieltges, A.; Großerüschkamp, F.; Mosig, A.; Diem, M.; Tannapfel, A.; Gerwert, K. Immunohistochemistry, histopathology and infrared spectral histopathology of colon cancer tissue sections. J. Biophotonics 2013, 6, 88–100. [Google Scholar] [CrossRef]
- Marzouk, O.; Schofield, J. Review of histopathological and molecular prognostic features in colorectal cancer. Cancers 2011, 3, 2767–2810. [Google Scholar] [CrossRef] [PubMed]
- Noothalapati, H.; Iwasaki, K.; Yamamoto, T. Non-invasive diagnosis of colorectal cancer by Raman spectroscopy: Recent developments in liquid biopsy and endoscopy approaches. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 258, 119818. [Google Scholar] [CrossRef]
- Kallaway, C.; Almond, L.M.; Barr, H.; Wood, J.; Hutchings, J.; Kendall, C.; Stone, N. Advances in the clinical application of Raman spectroscopy for cancer diagnostics. Photodiagn. Photodyn. Ther. 2013, 10, 207–219. [Google Scholar] [CrossRef]
- Dong, L.; Sun, X.; Chao, Z.; Zhang, S.; Zheng, J.; Gurung, R.; Du, J.; Shi, J.; Xu, Y.; Zhang, Y. Evaluation of FTIR spectroscopy as diagnostic tool for colorectal cancer using spectral analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 122, 288–294. [Google Scholar] [CrossRef]
- Kim, Y.I.; Jeong, S.; Jung, K.O.; Song, M.G.; Lee, C.H.; Chung, S.J.; Park, J.Y.; Cha, M.G.; Lee, S.G.; Jun, B.H.; et al. Simultaneous detection of EGFR and VEGF in colorectal cancer using fluorescence-Raman endoscopy. Sci. Rep. 2017, 7, 1035. [Google Scholar] [CrossRef] [PubMed]
- Petersen, D.; Naveed, P.; Ragheb, A.; Niedieker, D.; El-Mashtoly, S.F.; Brechmann, T.; Kötting, C.; Schmiegel, W.H.; Freier, E.; Pox, C.; et al. Raman fiber-optical method for colon cancer detection: Cross-validation and outlier identification approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 181, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Bergholt, M.S.; Lin, K.; Wang, J.; Zheng, W.; Xu, H.; Huang, Q.; Ren, J.-L.; Ho, K.Y.; The, M.; Srivastava, S.; et al. Simultaneous fingerprint and high-wavenumber fiber-optic Raman spectroscopy enhances real-time in vivo diagnosis of adenomatous polyps during colonoscopy. J. Biophotonics 2016, 9, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.A.; Jenkins, R.A.; Pryse, M.M.; Welsby, K.A.; Jitsumura, M.; Thornton, C.A.; Dunstan, P.R.; Harris, D.A. A high-throughput serum Raman spectroscopy platform and methodology for colorectal cancer diagnostics. Analyst 2018, 143, 6014–6024. [Google Scholar] [CrossRef]
- Synytsya, A.; Judexova, M.; Hoskovec, D.; Miskovicova, M.; Petruzelka, L. Raman spectroscopy at different excitation wavelengths (1064, 785 and 532 nm) as a tool for diagnosis of colon cancer. J. Raman Spectrosc. 2014, 45, 903–911. [Google Scholar] [CrossRef]
- Li, Q.B.; Xu, Z.; Zhang, N.W.; Zhang, L.; Wang, F.; Yang, L.M.; Wang, J.S.; Zhou, S.; Zhang, Y.F.; Zhou, X.S.; et al. In vivo and in situ detection of colorectal cancer using Fourier transform infrared spectroscopy. World J. Gastroenterol. 2005, 11, 327. [Google Scholar] [CrossRef]
- Li, S.; Chen, G.; Zhang, Y.; Guo, Z.; Liu, Z.; Xu, J.; Li, X.; Lin, L. Identification and characterization of colorectal cancer using Raman spectroscopy and feature selection techniques. Opt. Express 2014, 22, 25895–25908. [Google Scholar] [CrossRef]
- Zheng, F.; Chen, K.; Qin, Y. Sensitivity map of laser tweezers Raman spectroscopy for single-cell analysis of colorectal cancer. J. Biomed. Opt. 2007, 12, 034002. [Google Scholar] [CrossRef]
- Chen, K.; Qin, Y.; Zheng, F.; Sun, M.; Shi, D. Diagnosis of colorectal cancer using Raman spectroscopy of laser-trapped single living epithelial cells. Opt. Lett. 2006, 31, 2015–2017. [Google Scholar] [CrossRef]
- Yan, X.L.; Dong, R.X.; Zhang, L.; Zhang, X.J.; Zhang, Z.W. Raman spectra of single cell from gastrointestinal cancer patients. World J. Gastroenterol. 2005, 11, 3290. [Google Scholar] [CrossRef] [PubMed]
- Petersen, D.; Mavarani, L.; Niedieker, D.; Freier, E.; Tannapfel, A.; Kötting, C.; Gerwert, K.; El-Mashtoly, S.F. Virtual staining of colon cancer tissue by label-free Raman micro-spectroscopy. Analyst 2017, 142, 1207–1215. [Google Scholar] [CrossRef]
- Lloyd, G.R.; Wood, J.; Kendall, C.; Cook, T.; Shepherd, N.; Stone, N. Histological imaging of a human colon polyp sample using Raman spectroscopy and self organising maps. Vib. Spectrosc. 2012, 60, 43–49. [Google Scholar] [CrossRef]
- Liu, W.; Wang, H.; Du, J.; Jing, C. Raman microspectroscopy of nucleus and cytoplasm for human colon cancer diagnosis. Biosens. Bioelectron. 2017, 97, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, T.; Li, S.; Zhang, S.; Jin, L. Discrimination of rectal cancer through human serum using surface-enhanced Raman spectroscopy. Appl. Phys. B 2015, 119, 393–398. [Google Scholar] [CrossRef]
- Miskovicova, M.; Fryba, V.; Petruzelka, L.; Setnicka, V.; Synytsya, A.; Tatarkovic, M.; Ulrych, J.; Vocka, M. Novel spectroscopic biomarkers are applicable in non-invasive early detection and staging classification of colorectal cancer. Neoplasma 2020, 67, 1349–1358. [Google Scholar] [CrossRef]
- Tatarkovič, M.; Miškovičová, M.; Šťovíčková, L.; Synytsya, A.; Petruželka, L.; Setnička, V. The potential of chiroptical and vibrational spectroscopy of blood plasma for the discrimination between colon cancer patients and the control group. Analyst 2015, 140, 2287–2293. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, D.; Lin, J.; Yu, Y.; Huang, Z.; Chen, Y.; Lin, J.; Feng, S.; Li, B.; Liu, N.; et al. Label-free detection of serum proteins using surface-enhanced Raman spectroscopy for colorectal cancer screening. J. Biomed. Opt. 2014, 19, 087003. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Judexová, M.; Hrubý, T.; Tatarkovič, M.; Miškovičová, M.; Petruželka, L.; Setnička, V. Analysis of human blood plasma and hen egg white by chiroptical spectroscopic methods. Anal. Bioanal. Chem. 2013, 405, 5441–5453. [Google Scholar] [CrossRef]
- Liu, W.; Sun, Z.; Chen, J.; Jing, C. Raman spectroscopy in colorectal cancer diagnostics: Comparison of PCA-LDA and PLS-DA models. J. Spectrosc. 2016, 2016, 1603609. [Google Scholar] [CrossRef]
- Choi, J.S.; Kim, J.; Hong, Y.J.; Bae, W.Y.; Choi, E.H.; Jeong, J.W.; Park, H.K. Evaluation of non-thermal plasma-induced anticancer effects on human colon cancer cells. Biomed. Opt. Express 2017, 8, 2649–2659. [Google Scholar] [CrossRef]
- Kaznowska, E.; Depciuch, J.; Szmuc, K.; Cebulski, J. Use of FTIR spectroscopy and PCA-LDC analysis to identify cancerous lesions within the human colon. J. Pharm. Biomed. Anal. 2017, 134, 259–268. [Google Scholar] [CrossRef]
- Li, X.; Yang, T.; Li, S.; Wang, D.; Song, Y.; Zhang, S. Raman spectroscopy combined with principal component analysis and k nearest neighbour analysis for non-invasive detection of colon cancer. Laser Phys. 2016, 26, 035702. [Google Scholar] [CrossRef]
- Khanmohammadi, M.; Garmarudi, A.B.; Samani, S.; Ghasemi, K.; Ashuri, A. Application of linear discriminant analysis and attenuated total reflectance Fourier transform infrared microspectroscopy for diagnosis of colon cancer. Pathol. Oncol. Res. 2011, 17, 435–441. [Google Scholar] [CrossRef]
- Molckovsky, A.; Song, L.M.W.K.; Shim, M.G.; Marcon, N.E.; Wilson, B.C. Diagnostic potential of near-infrared Raman spectroscopy in the colon: Differentiating adenomatous from hyperplastic polyps. Gastrointest. Endosc. 2003, 57, 396–402. [Google Scholar] [CrossRef]
- Manavbasi, Y.; Süleymanoglu, E. Nucleic acid-phospholipid recognition: Fourier transform infrared spectrometric characterization of ternary phospho-lipid-inorganic cation-DNA complex and its relevance to chemicopharmaceutical design of nanometric liposome based gene delivery formulations. Arch. Pharm. Res. 2007, 30, 1027–1040. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, D.I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Yao, H.; Shi, X.; Zhang, Y. The use of FTIR-ATR spectrometry for evaluation of surgical resection margin in colorectal cancer: A pilot study of 56 samples. J. Spectrosc. 2014, 2014, 213890. [Google Scholar] [CrossRef]
- Ramesh, J.; Salman, A.; Mordechai, S.; Argov, S.; Goldstein, J.; Sinelnikov, I.; Walfish, S.; Guterman, H. FTIR microscopic studies on normal, polyp, and malignant human colonic tissues. Subsurf. Sens. Technol. Appl. 2001, 2, 99–117. [Google Scholar] [CrossRef]
- Barth, A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 2000, 74, 141–173. [Google Scholar] [CrossRef]
- Kondepati, V.R.; Heise, H.M.; Oszinda, T.; Mueller, R.; Keese, M.; Backhaus, J. Detection of structural disorders in colorectal cancer DNA with Fourier-transform infrared spectroscopy. Vib. Spectrosc. 2008, 46, 150–157. [Google Scholar] [CrossRef]
- Olsztyńska-Janus, S.; Komorowska, M. Conformational changes of L-phenylalanine induced by near infrared radiation. ATR-FTIR studies. Struct. Chem. 2012, 23, 1399–1407. [Google Scholar] [CrossRef]
- Olsztynska, S.; Komorowska, M.; Vrielynck, L.; Dupuy, N. Vibrational spectroscopic study of L-phenylalanine: Effect of pH. Appl. Spectrosc. 2001, 55, 901–907. [Google Scholar] [CrossRef]
- Hernández, B.; Pflüger, F.; Adenier, A.; Kruglik, S.G.; Ghomi, M. Vibrational analysis of amino acids and short peptides in hydrated media. VIII. Amino acids with aromatic side chains: L-phenylalanine, L-tyrosine, and L-tryptophan. J. Phys. Chem. B 2010, 114, 15319–15330. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Beattie, J.R.; Bell, S.E.; Moss, B.W. A critical evaluation of Raman spectroscopy for the analysis of lipids: Fatty acid methyl esters. Lipids 2004, 39, 407–419. [Google Scholar] [CrossRef]
- Krafft, C.; Ramoji, A.A.; Bielecki, C.; Vogler, N.; Meyer, T.; Akimov, D.; Rösch, P.; Schmitt, M.; Dietzek, B.; Petersen, I.; et al. A comparative Raman and CARS imaging study of colon tissue. J. Biophotonics 2009, 2, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R.; Caspers, P.; Puppels, G.J.; Pandiancherri, S.; McNaughton, D. Resonance Raman spectroscopy of red blood cells using near-infrared laser excitation. Anal. Bioanal. Chem. 2007, 387, 1691–1703. [Google Scholar] [CrossRef]
- Hu, S.; Smith, K.M.; Spiro, T.G. Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 1996, 118, 12638–12646. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Treiman, M.; Brazhe, A.R.; Find, N.L.; Maksimov, G.V.; Sosnovtseva, O.V. Mapping of redox state of mitochondrial cytochromes in live cardiomyocytes using Raman microspectroscopy. PLoS ONE 2012, 7, e41990. [Google Scholar] [CrossRef]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D.L. The Mahalanobis distance. Chemom. Intell. Lab. Syst. 2000, 50, 1–18. [Google Scholar] [CrossRef]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Scalfi-Happ, C.; Udart, M.; Hauser, C.; Rück, A. Investigation of lipid bodies in a colon carcinoma cell line by confocal Raman microscopy. Med. Laser Appl. 2011, 26, 152–157. [Google Scholar] [CrossRef]
- Accioly, M.T.; Pacheco, P.; Maya-Monteiro, C.M.; Carrossini, N.; Robbs, B.K.; Oliveira, S.S.; Kaufmann, K.; Morgado-Diaz, J.A.; Bozza, P.T.; Viola, J.P. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res. 2008, 68, 1732–1740. [Google Scholar] [CrossRef]
- Kurabe, N.; Hayasaka, T.; Ogawa, M.; Masaki, N.; Ide, Y.; Waki, M.; Nakamura, T.; Kurachi, K.; Kahyo, T.; Shinmura, K.; et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT 4. Cancer Sci. 2013, 104, 1295–1302. [Google Scholar] [CrossRef]
- Lavezzi, A.M.; Ottaviani, G.; De Ruberto, F.; Fichera, G.; Matturri, L. Prognostic significance of different biomarkers (DNA content, PCNA, karyotype) in colorectal adenomas. Anticancer Res. 2002, 22, 2077–2081. [Google Scholar]
- Jin, W.; Gao, M.Q.; Lin, Z.W.; Yang, D.X. Quantitative study of multiple biomarkers of colorectal tumor with diagnostic discrimination model. World J. Gastroenterol. 2004, 10, 439. [Google Scholar] [CrossRef]
- Birk, J.W.; Tadros, M.; Moezardalan, K.; Nadyarnykh, O.; Forouhar, F.; Anderson, J.; Campagnola, P. Second harmonic generation imaging distinguishes both high-grade dysplasia and cancer from normal colonic mucosa. Dig. Dis. Sci. 2014, 59, 1529–1534. [Google Scholar] [CrossRef]
- Kirkland, S.C. Type I collagen inhibits differentiation and promotes a stem cell-like phenotype in human colorectal carcinoma cells. Br. J. Cancer 2009, 101, 320–326. [Google Scholar] [CrossRef]
- Zou, X.; Feng, B.; Dong, T.; Yan, G.; Tan, B.; Shen, H.; Huang, A.; Zhang, X.; Zhang, M.; Yang, P.; et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. J. Proteom. 2013, 94, 473–485. [Google Scholar] [CrossRef]
- Moilanen, J.M.; Kokkonen, N.; Löffek, S.; Väyrynen, J.P.; Syväniemi, E.; Hurskainen, T.; Mäkinen, M.; Klintrup, K.; Mäkelä, J.; Sormunen, R.; et al. Collagen XVII expression correlates with the invasion and metastasis of colorectal cancer. Hum. Pathol. 2015, 46, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Fenyvesi, A. The prognostic significance of type IV collagen expression in colorectal carcinomas. Arch. Oncol. 2003, 11, 65–70. [Google Scholar] [CrossRef][Green Version]
- Mohamed, H.T.; Untereiner, V.; Sockalingum, G.D.; Brézillon, S. Implementation of infrared and Raman modalities for glycosaminoglycan characterization in complex systems. Glycoconj. J. 2017, 34, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Brézillon, S.; Untereiner, V.; Lovergne, L.; Tadeo, I.; Noguera, R.; Maquart, F.X.; Wegrowski, Y.; Sockalingum, G.D. Glycosaminoglycan profiling in different cell types using infrared spectroscopy and imaging. Anal. Bioanal. Chem. 2014, 406, 5795–5803. [Google Scholar] [CrossRef]
- Mainreck, N.; Brézillon, S.; Sockalingum, G.D.; Maquart, F.-X.; Manfait, M.; Wegrowski, Y. Characterization of glycosaminoglycans by tandem vibrational microspectroscopy and multivariate data analysis. Methods Mol. Biol. 2012, 836, 117–130. [Google Scholar]
- Mainreck, N.; Brézillon, S.; Sockalingum, G.D.; Maquart, F.X.; Manfait, M.; Wegrowski, Y. Rapid characterization of glycosaminoglycans using a combined approach by infrared and Raman microspectroscopies. J. Pharm. Sci. 2011, 100, 441–450. [Google Scholar] [CrossRef]
- Kalathas, D.; Theocharis, D.A.; Bounias, D.; Kyriakopoulou, D.; Papageorgakopoulou, N.; Stavropoulos, M.S.; Vynios, D.H. Chondroitin synthases I, II, III and chondroitin sulfate glucuronyltransferase expression in colorectal cancer. Mol. Med. Rep. 2011, 4, 363–368. [Google Scholar] [PubMed]
- Brauchle, E.; Kasper, J.; Daum, R.; Schierbaum, N.; Falch, C.; Kirschniak, A.; Schäffer, T.E.; Schenke-Layland, K. Biomechanical and biomolecular characterization of extracellular matrix structures in human colon carcinomas. Matrix Biol. 2018, 68, 180–193. [Google Scholar] [CrossRef]
- Kalathas, D.; Theocharis, D.A.; Bounias, D.; Kyriakopoulou, D.; Papageorgakopoulou, N.; Stavropoulos, M.S.; Vynios, D.H. Alterations of glycosaminoglycan disaccharide content and composition in colorectal cancer: Structural and expressional studies. Oncol. Rep. 2009, 22, 369–375. [Google Scholar]
- Laneza, A.; Vizoso, F.; Rodriguez, J.C.; Raigoso, P.; Garcia-Muniz, J.L.; Allende, M.T.; Garcia-Moran, M. Hyaluronic acid as prognostic marker in resectable colorectal cancer. Br. J. Surg. 2000, 87, 1690–1696. [Google Scholar] [CrossRef]
- Wu, R.L.; Huang, L.; Zhao, H.C.; Geng, X.P. Hyaluronic acid in digestive cancers. J. Cancer Res. Clin. Oncol. 2017, 143, 1–16. [Google Scholar] [CrossRef]
- Annibaldi, A.; Widmann, C. Glucose metabolism in cancer cells. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Sakr, S.A.; Abdel-Wahed, M.M.; Abdou, A.G.; El-Adely, E.K. Histochemical alterations in colorectal carcinoma and adenoma in Egyptian patients. J. Coast. Life Med. 2016, 4, 14–20. [Google Scholar] [CrossRef]

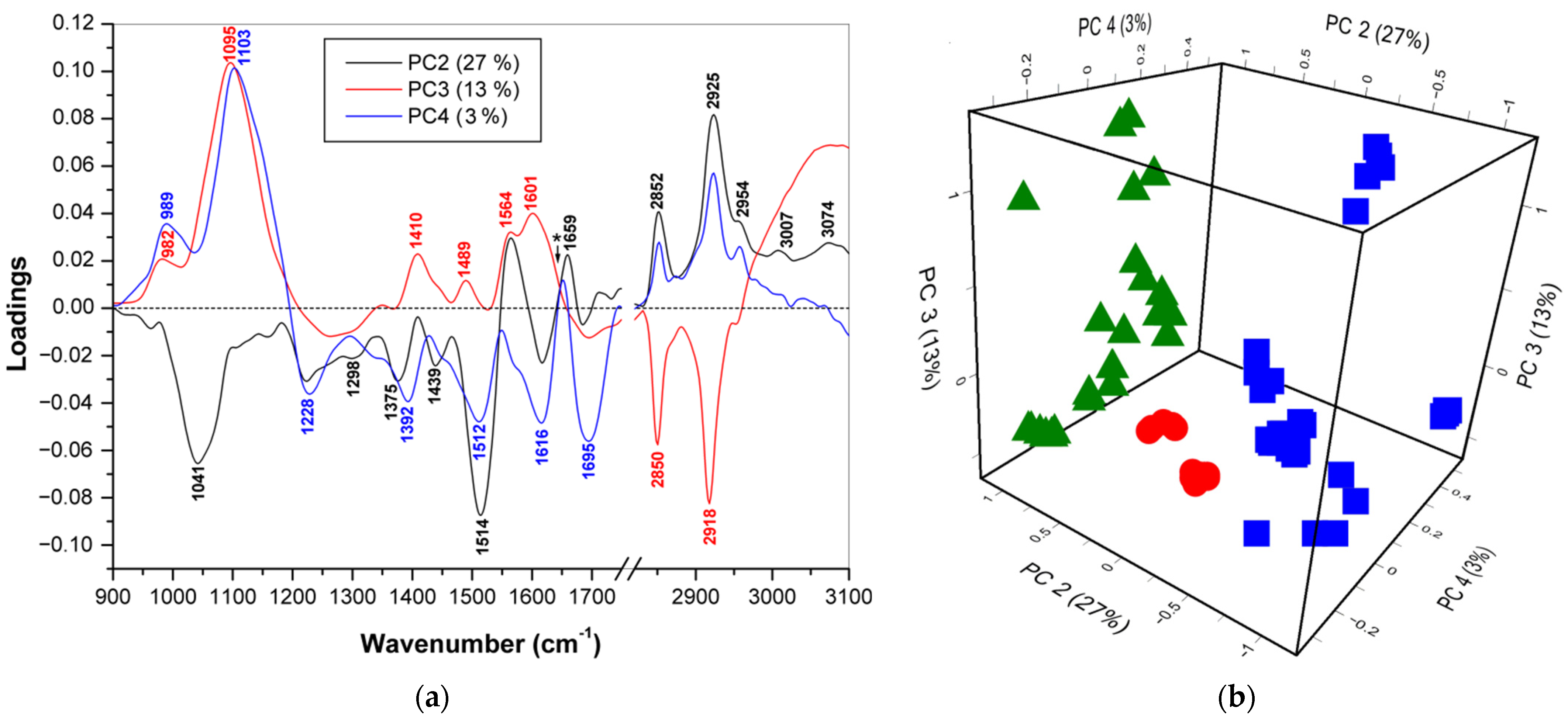


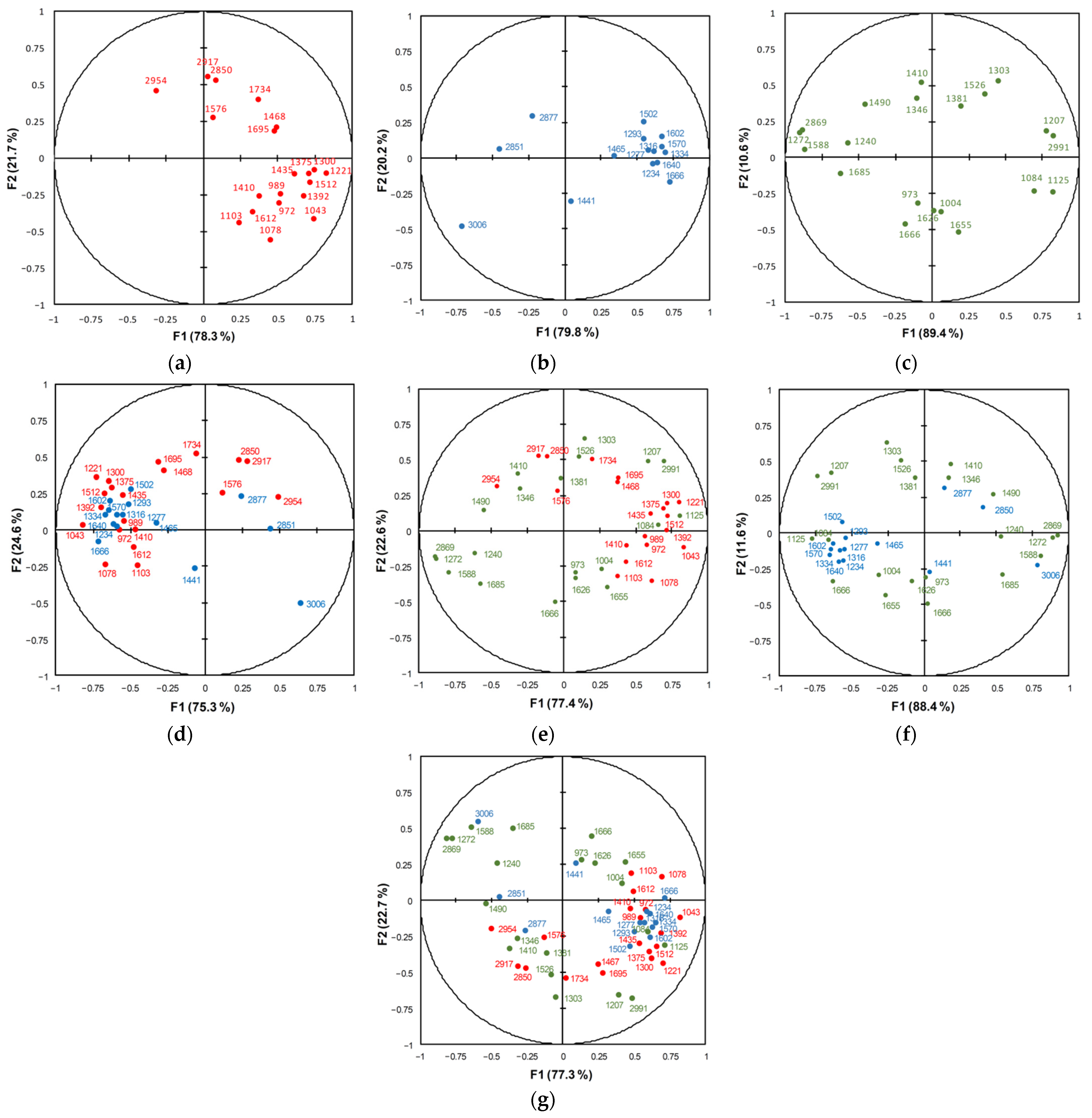
| Method | Variables: Wavenumber (cm−1) |
|---|---|
| FTIR | 972, 989, 1043, 1078, 1103, 1221, 1300, 1375, 1392, 1410, 1435, 1468, 1512, 1576, 1612, 1695, 1734, 2850, 2917, 2954 |
| FT Raman | 1234, 1277, 1293, 1316, 1334, 1441, 1465, 1502, 1570, 1602, 1640, 1666, 2851, 3006 |
| Dispersion Raman | 973, 1004, 1084, 1125, 1207, 1240, 1272, 1303, 1346, 1381, 1410, 1490, 1526, 1588, 1626, 1655, 1666, 1685, 2869, 2991 |
| Dataset | Distances between the Diagnostic Groups (Mean ± SD) | ||
|---|---|---|---|
| n ↔ a | n ↔ c | a ↔ c | |
| FTIR | 959.2 ± 43.5 | 1048.2 ± 39.4 | 329.0 ± 25.8 |
| FT Raman | 329.0 ± 17.4 | 83.6 ± 16.4 | 63.8 ± 16.4 |
| Dispersion Raman | 314.9 ± 31.9 | 360.5 ± 33.5 | 88.3 ± 24.6 |
| FTIR + FT Raman | 2859.5 ± 69.8 | 1944.4 ± 65.8 | 1793.5 ± 65.0 |
| FTIR + Dispersion Raman | 3243.1 ± 79.0 | 2548 ± 70.2 | 1648.0 ± 68.6 |
| FT Raman + Disp. Raman | 2243.3 ± 72.0 | 1965.1 ± 73.4 | 516.9 ± 61.5 |
| FTIR + FT Raman + Disp. Raman | 8127.4 ± 146.6 | 4798.5 ± 118.3 | 5718.0 ± 102.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Synytsya, A.; Vaňková, A.; Miškovičová, M.; Petrtýl, J.; Petruželka, L. Ex Vivo Vibration Spectroscopic Analysis of Colorectal Polyps for the Early Diagnosis of Colorectal Carcinoma. Diagnostics 2021, 11, 2048. https://doi.org/10.3390/diagnostics11112048
Synytsya A, Vaňková A, Miškovičová M, Petrtýl J, Petruželka L. Ex Vivo Vibration Spectroscopic Analysis of Colorectal Polyps for the Early Diagnosis of Colorectal Carcinoma. Diagnostics. 2021; 11(11):2048. https://doi.org/10.3390/diagnostics11112048
Chicago/Turabian StyleSynytsya, Alla, Aneta Vaňková, Michaela Miškovičová, Jaromír Petrtýl, and Luboš Petruželka. 2021. "Ex Vivo Vibration Spectroscopic Analysis of Colorectal Polyps for the Early Diagnosis of Colorectal Carcinoma" Diagnostics 11, no. 11: 2048. https://doi.org/10.3390/diagnostics11112048
APA StyleSynytsya, A., Vaňková, A., Miškovičová, M., Petrtýl, J., & Petruželka, L. (2021). Ex Vivo Vibration Spectroscopic Analysis of Colorectal Polyps for the Early Diagnosis of Colorectal Carcinoma. Diagnostics, 11(11), 2048. https://doi.org/10.3390/diagnostics11112048






