1. Introduction
When a pandemic spreads rapidly and widely around the world, developing a mathematical model becomes a useful tool to investigate the impact and spread of the viruses. Numerous models have been proposed in the literature to predict the spread of the diseases in order to come out with better disease control policies and measures. To describe the spread of the disease, techniques and ideas from different fields of study have been applied. For example, Altan and Kaasu [
1] proposed a model consisting of a two-dimensional curvelet transformation, a met-heuristic optimization algorithm, and a deep learning technique to diagnose the infected patient from X-ray images. Balli [
2] used a machine learning time series prediction model to obtain the disease curve and forecast the epidemic trend, while Bassey and Atsu [
3] considered the model using dual-bilinear control treatment functions arising from dual non-pharmaceutical controls and dual pharmaceutical therapies to compute the system reproduction numbers and to analyze the system stability. Butt et al. [
4] and Hanif et al. [
5] proposed the Caputo–Fabrizio-fractional model to study the dynamical behavior of the pandemic and to prove the existence and uniqueness of the solutions using fractional calculus and a functional analysis, and so on.
Among the various models, the compartmental models are a very useful and common modelling technique. The most frequently investigated mathematical model of epidemics is the susceptible (S)–infected (I)–recovered (R) (SIR) model. It is a deterministic epidemic model in which the individuals are classified into three individuals that can only move from S to I and from I to R. Afterwards, many studies were conducted by generalizing this simplest compartment model. For example, Wu et al. [
6] used the susceptible–exposed–infected–recovered metapopulation model (the SEIR model) to simulate epidemics and estimated the basic reproduction number via Markov Chain Monte Carlo methods, while Khajanchi and Sarkar [
7] proposed a mathematical model called the SAIUQR model with six compartments, namely, susceptible (S), asymptomatic (A), reported symptomatic (I), unreported symptomatic (U), quarantine (Q), and recovered (R), in order to study the feasible equilibria and the stability with respect to the basic reproduction number, etc. However, it has been noticed that many compartment models are valid only in the case of sufficiently large populations, and hence, some stochastic models are presented. Britton [
8] defined the standard stochastic SIR epidemic model for a closed population in order to study small population properties and a large population approximation. Bittihn and Golestanian [
9] used a stochastic SIR metapopulation model for disease transmission and divided this large population into some sub-populations in which the number of infected individuals was low to estimate the effects of extinction and desynchronization. Faranda and Alberti [
10] used the stochastic SEIR model in which the incubation, infection, and recovery rates are stochastically perturbed to investigate the long-term behavior of the epidemic evolution. He et al. [
11] developed a discrete-time stochastic epidemic model with binomial distribution to forecast the spread of the disease in the next period and to evaluate the risk in order to provide suggestions on the returning timing to the routine. Khan et al. [
12] used the stochastic Volterra integro-differential equation to model the disease transmission and used the spectral collocation method to approximate the solution of the equation.
In particular, multitype branching processes provide another stochastic approach to model the spread of the disease. Jacob [
13] introduced various settings of branching processes for modeling the epidemic. According to the aims of the study, branching processes with different offspring distributions, initial conditions, or parameters are chosen to fit the population evolution. Britton [
8] constructed the stochastic SIR model using the branching process with the exponential distributed infected period for an approximation of the early stage of an outbreak. Hellewell et al. [
14] considered a branching process with a negative binomial offspring distribution to measure the success of controlling outbreaks using isolation and contact tracing. Levesque et al. [
15] used a continuous time branching processes with a negative binomial offspring distribution to develop the efficacy of contact tracing strategies. Laha [
16] considered a time-varying multitype branching process model for the spread of infectious disease in which it is assumed that all detected individuals are completely contact traced and the expected numbers of detected infected individuals, undetected infected individuals, and the total number of cases up to generation
n are computed. Laha and Majumdar [
17] extended the multitype branching model to the case in which the contact tracing is only partial and used Poisson offspring distributions whose parameters may vary over time to investigate the expected number of individuals in each category. Yanev et al. [
18] considered both two-type (infected undetected vs infected detected) branching processes with or without immigration and studied the growth rate and so on.
In this article, we introduced a stochastic model using the multitype branching processes with a general offspring distribution for which the offspring mean matrix is no longer primitive but can be broken down into two primitive components. We investigated the asymptotic growth rate of the population vector and the spread rate of each type in an infectious disease according to the magnitude of the maximal eigenvalues of these two component matrices of the offspring mean matrix.
This paper is organized as follows. In
Section 2, we introduce the setting of the stochastic spread model using branching processes. In
Section 3, we present the results on the asymptotic growth rate, the properties of the population vector, and the spread rate of each type. Finally, the proofs of the main results are provided in
Section 4.
2. Random Spread Models Using Branching Processes
To introduce a spread model using the multitype branching processes, we considered a population in which individuals are classified into
K types, say
, where individuals in different categories can be considered as patients in different categories such as highly contagious, moderately contagious, mildly contagious, and so on. We assume that this population starts with one individual, i.e., the spread of the disease initiated by one individual at time 0 (or in the 0th generation). During the pandemic, the viruses are passed from person to person. We call an individual who is infected a “child” or an offspring of the individual who passes the viruses on, while the one passing the viruses on is called the “parent”. When the infected population evolves over time, we are interested only in the number of “children” each parent has. So, we assume that each individual lives (the time when this individual is able to infect others) a unit of time and, upon its death (recovery), produces its offspring according to the probability distribution
, where
is the probability that an individual of type
produces
children of type
,
children of type
, ⋯, and
children of type
. This reproduction is also assumed to be independent of others in the same generation and in the past of the population. Let
be the population vector in the
nth generation (or at time
n), where
is the number of individuals of type
in the
nth generation,
. Then, the process
is called a
K-type branching process with offspring distribution
.
Let
be the expected value of the number of children of type
produced by an individual of type
, where
is the unit vector with 1 as its
ith component. Then, the matrix
is called the
offspring mean matrix for this branching process
. Moreover, if
and
for all
; then, the
-entry
of the matrix
is the expected value
of the number of offspring of type
in the
nth generation of the population initiated by an ancestor of type
.
We know from the classical theory of branching processes that the behavior of the offspring matrix provides information about the branching process in the long run. In particular, the classical limit theorem tells that, when the branching process is non-singular and the offspring mean matrix
is primitive with a maximal eigenvalue
, under some proper conditions, the population vector grows geometrically almost surely:
where
is the normalized left eigenvector associated with
and
W is a random variable. We refer readers to Athreya and Ney [
19] for more details.
To analyze the population composition in the long run, we define the spread rate of each type
as the limit, if it exists in some sense, of the proportion of individuals of type
in the
nth generation of the population initiated with one individual of type
as
. Namely, conditioned on the event of
,
where
. The geometric convergence of the population vector stated in (
1) above is usually the key to studying the spread rate of each type. However, in some situations, the offspring mean matrix is no longer primitive. For example, the spread model with frozen symbols proposed by Ban et al. [
20] forms the non-primitive case. So, in this paper, we focus on the stochastic spread model with a non-primitive offspring mean matrix.
In addition, we define the covariance matrix
as the following:
where its
-entry is
for all
. For each
, we also define the matrix
in which the
-entry is
for all
. Note that this matrix
is symmetric, and the only nonzero entry in
is the
-entry. According to Harris [
21] (p. 37),
can be written in the following form:
With the proper condition of the covariance matrices , we can show the geometric growth rate of , which leads to the geometric growth of the population vector.
3. Main Results on the Growth and Spread Rates
Throughout this paper, the branching process is assumed to be non-singular and the offspring mean matrix is assumed to be of the following form:
where the
matrix
A and the
matrix
B are primitive and
C is a non-zero matrix. Then, by induction and computation, we can find the form of
:
As shown in Ban et al. [
20], there exists an
N such that
.
We adapt the following notations: for any square matrix
M,
denotes the maximal eigenvalue of
M, and
and
denote the right and left eigenvectors of
M associated with
, respectively. In particular, if
is the offspring mean matrix, we write
for
,
for
and
for
, respectively. We also normalize
and
such that
The first theorem in this section reveals the geometric growth of the population vector:
Theorem 1. If and the covariance matrices ’s are finite for all , then there exists a random vector with such thatboth in mean square and with probability 1. The next theorem tells us the property of the limit vector . Namely, it has the same direction as the left eigenvector of the offspring mean matrix .
Theorem 2. If , the covariance matrices are finite for all and the random vector is defined as in Theorem 1. Then, there exists a random variable W such thatwith probability 1. Moreover, for all . Remark 1. The left eigenvector of the offspring matrix associated with the maximal eigenvalue ρ, where and , has the following properties:
- (i)
If , then , and is a left eigenvector associated with ;
- (ii)
If , then , and is a left eigenvector associated with .
Similar results also hold for the right eigenvector of the offspring matrix associated with ρ, where and :
- (i)
If , then , and is a right eigenvector associated with ;
- (ii)
If , then , and is a right eigenvector associated with .
We refer the readers to Ban et al. [20] for more details. As a consequence of Theorems 1 and 2, we obtain the spread rate for the random spread model in the next two theorems according to the magnitude of and .
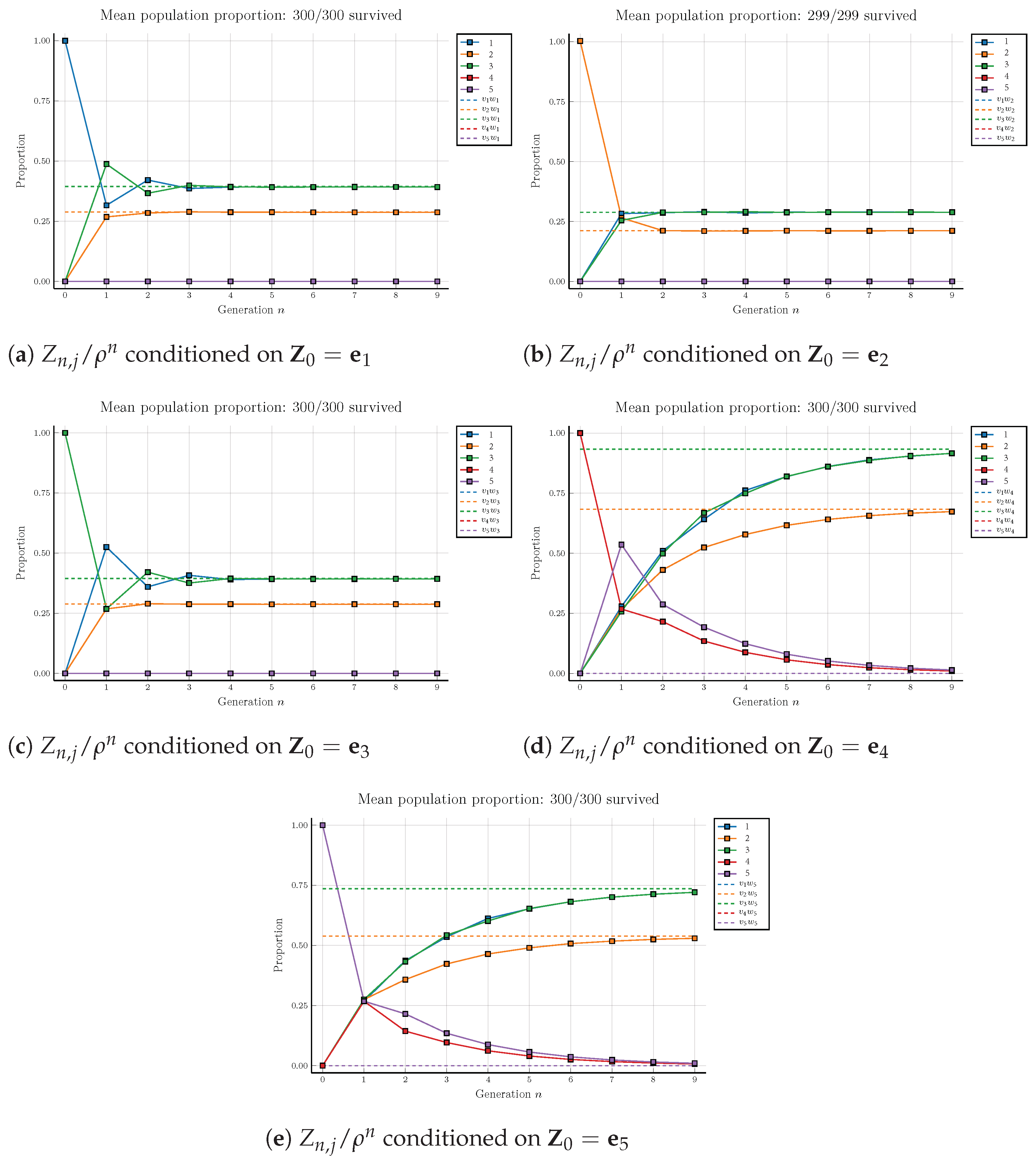
Theorem 3. Let and the covariance matrices be finite, and let . Then, in the event , the spread ratewith probability 1, where , is the normalized left eigenvector of A associated with the maximal eigenvalue . Theorem 4. Let and the covariance matrices be finite and let . Then, in the event , the spread ratewith probability 1. 6. Discussion and Conclusions
To model the random phenomenon of the spread of the disease during a pandemic, we propose a spread model using the multitype branching process. Due to the discreteness of the processes, we are able to study the spread pattern at any time
n by studying the behavior of the
nth power of the offspring mean matrix. However, in order to be able to consider more applications which are not supported by the classical results in the theory of the branching processes due to the non-primitive offspring mean matrix, we consider the non-primitive offspring mean matrix which is of the form
with primitive components
A and
B and positive component
C and study the asymptotic growth rate of the population vector and the spread rate of each type in the long run. In this work, we prove that the population vector grows geometrically (Theorem 1) and the limit of the normalized population vector share the same direction as the left eigenvector of the offspring mean associated with the maximal eigenvalue (Theorem 2). Moreover, we show that the limit of the type composition is proportional to the left eigenvector associated with the maximal eigenvalue of the offspring mean matrix and find the spread rates (Theorems 3 and 4). Although we only state the results and provide the proofs for the cases when the offspring mean matrix has two primitive components, similar discussions should be adapted to establish the spread rate for even more general forms such as
with primitive components
,
and the results could be achieved under weaker conditions on the moment conditions of the offspring matrix.