1. Introduction
Every part of our lives is being impacted by digitalization. Therefore, data analytics systems provide enormous advantages in healthcare industries [
1,
2] and other sectors, such as security [
3], text processing [
4], and finance [
5]. Modern medical systems produce enormous amounts of data every day. To extract valuable information and identify hidden patterns in these data, mining and analysis are required [
6]. The benefits of data analytics systems in the healthcare sector extend from disease investigation to treatment levels to analyzing the pathology report, which provides an interpretation of the results from the patient’s body sample, and these reports are considered undoubtedly one of the most critical documents in medicine [
7]. Typically, pathology reports include various pieces of essential information about the patient’s symptomatology. These reports are in free-text semi-structured or unstructured formats. The data analysts in this scenario manually examine the pathology reports, extract useful information, and then interpret the information in compliance with diagnostic features [
8]. Finally, the results are finalized and entered into the database based on several computer processes [
9]. Text mining has developed as a practical computer tool to accurately translate pathology reports into a usable structured representation by extracting only the important information that affects the hematological disease. For example, a string matching algorithm may provide helpful tools for data analysis [
10]. Therefore, it is evident that any new technology that can automatically handle pathology report data will become an area of interest. So, this paper is used to consolidate and characterize the use of CBC-driven parameters using a hybrid algorithm. Therefore, it can be concluded that many challenges can be countered with the application of data analytics technology in healthcare [
11]:
Data Quality;
Data Variety;
Data Validity, meaning the suitability of the data for their intended application;
Data Security;
Data Storage because of the large amount of data supplied;
Data Visualization may be needed in some cases;
Healthcare Data should be updated frequently to remain up-to-date and valuable.
Unlike previous studies, this paper proposes an efficient, dedicated algorithm for string matching to be used for the pathological analysis process to override the limitations in the best-chosen algorithms specified in the implementation of the comparisons study step. The main contributions are identified as follows:
This paper delves thoroughly into the probability of encountering many challenges in the application of data analytics technology to health care.
Several studies have presented the possibilities of using data analytics in healthcare application sectors.
This paper also identifies the most powerful string matching methods that can be used in healthcare by performing a comparative study of these methods.
Moreover, an enhanced Rapin–Karp algorithm was presented by integrating the Rapin–Karp algorithm after several modifications to find the exact matching with the possibility that there are no accurate words matched. The fuzzy ratio method was used to find the approximate matching.
The paper is organized as follows:
Section 2 provides a brief literature review of the studies on healthcare test analyses and string matching algorithms that would help the analyzing systems.
Section 3 presents the proposed system methodology, while
Section 4 interprets and describes the significance of the results of the proposed method. Finally,
Section 5 and
Section 6 provide a compelling discussion and conclusions that inform researchers on what they can learn from the research results.
3. The Proposed System Methodology
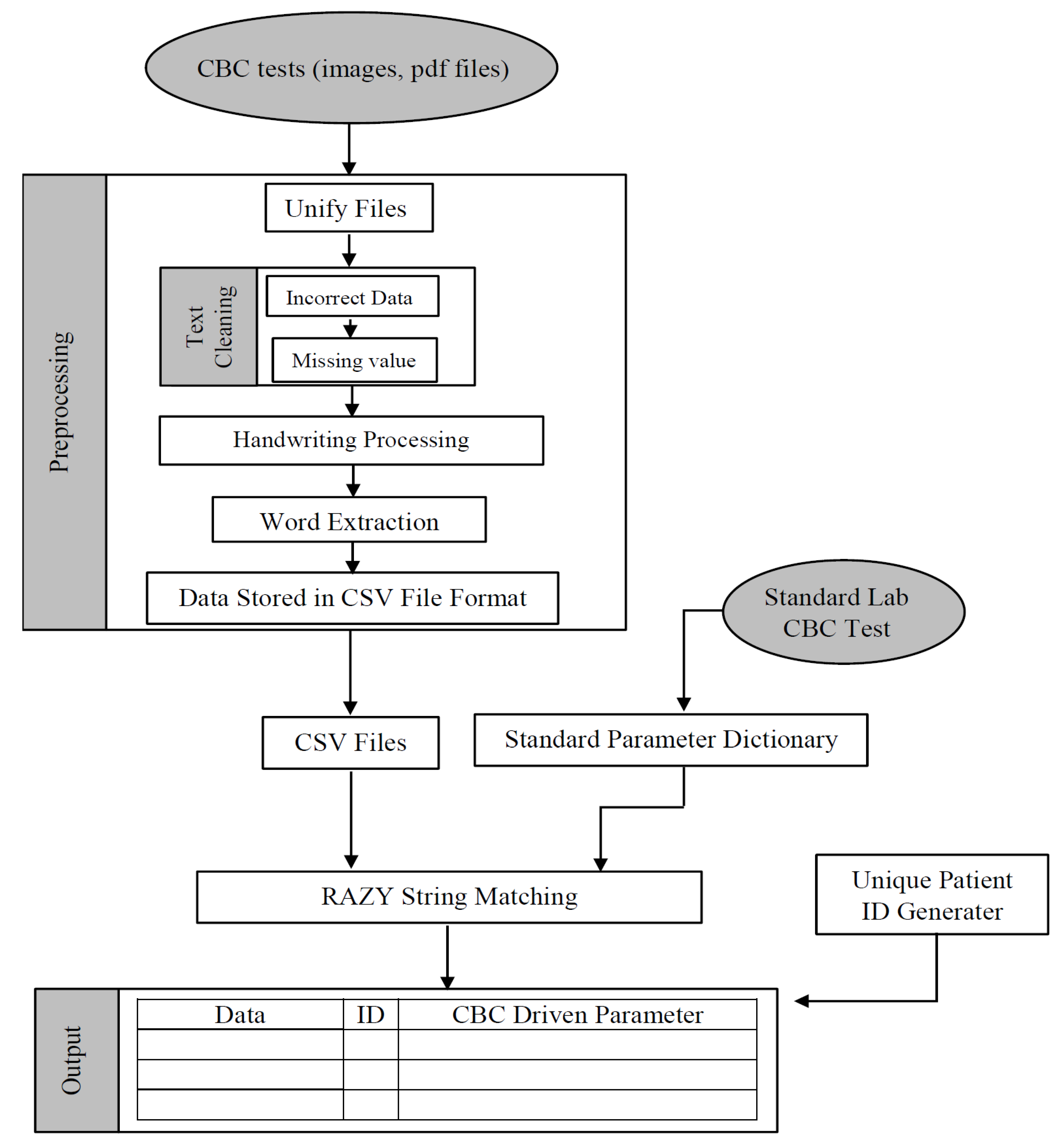
This methodology is for the creation of the general patient report (comprehensive prescription information). The method entails several steps, as shown in
Figure 1, which describes the proposed workflow of the proposed system.
3.1. Research Setting and Data Collection
In this paper, the CBC reports were collected as research symbols from two main places: the laboratory in the Al-Zahra hospital and the Hematology Center in the Medical City in Baghdad, Iraq. First, the data are aggregated from each patient as an initial step toward building a unified health information system to computerize patient records and create a general report for each patient. Then, as testing data, 150 of the most common terms (e.g., WBC, RBC, …), with their numerical values, used in the reports were selected. Next, the raw data were aggregated and comprised the individual patient level. They were then extracted to add to the patient records. In the case of adding a patient for the first time, a new record is created and generates a unique code as a patient identifier, i.e., the patient ID consists of 12 digits (for example, XXXX-XXXX-XXXX), and the content of this ID is as follows:
The first pair indicates the computer number (device) that was used to add the patient ID;
Two digits refer to the year (its value is the time of the ID’s addition);
Two digits refer to the month (its value is the time of the ID’s addition);
Two digits refer to the day (its value is the time of the ID’s addition);
Two digits refer to the hour (its value is the time of the ID’s addition);
Two digits refer to the seconds (its value is the time of the ID’s addition).
3.2. Automated Text Processing
Reading Pathological Reports: The initial step is performed depending on the current values extracted from the file (test name and test values) to facilitate data collection to update the patient’s history based on the proposed method. It is important to note that the reports may be images or PDF files. Therefore, we should unify the reading and analyzing of texts from the pathological reports process because each file type’s reading process differs. So, the reading process might affect the extracted text, and the possibility of an error occurring increases. Therefore, this step is divided into two parts. Firstly, convert all pdf files to image files (here, all files are converted to images). Then, extract the text from each image using the Tesseract-OCR engine (developed by Hewlett-Packard [
24]). After extracting the data, the progressive stages of the proposed system will be implemented to update the patient’s information based on the extracted values.
Data Cleaning: When text is extracted from a file, irrelevant symbols such as “*”, “;” …, and so on can be found. As a result, the text is processed to remove irrelevant information, extract only the most essential words and their values, and export this to a CSV file for further processing.
Error Handling: There is a possibility of existing handwritten (terms or values) inside the pathological reports. Therefore, the system should be trained to read files and ensure that the reading process is completed correctly for these terms. Moreover, the system should look at all of the words in the text to identify familiar (standard) and unfamiliar (wrong) words that could be misspelled. A similarity metrics technique, such as the Jaro–Winkler, dice coefficient, matching coefficient, or overlap coefficient) [
25] or distance metrics technique (such as the Levenshtein, Damerau–Levenshtein, longest common subsequence, or N-Gram) [
26] should be used to handle words that are unusual in pathological reports. In the case that an error occurs in the analysis terms, for example, the white blood cell rate analysis (e.g., WBD instead of WBC), there are two types of solutions, as shown below:
- –
They are adopting the modified string matching algorithm and the word dictionary method (which contains the common pathological words) to identify the similarity percentage and adopt the word with the most nominal rate of difference depending on the pre-specified threshold. The laboratory specialist should pre-specify the threshold and assume the risk of using the wrong analysis terms due to the importance of this information.
- –
Suppose the specified threshold value is not met. In this case, the system should show a message alerting the presence of an ambiguous analysis term in the pathological report that was not identified by standard methods and present the closest term.
Suppose there are missing values in the pathological report. In that case, the system can adopt the values found in the previous reports with a time limit (e.g., should not exceed three months) to maintain the shift-life time of information. If the missing values are unavailable within this time limit, a message is displayed in the system describing the situation (i.e., the findings acquired and the date of acquisition).
3.3. Best Algorithms Selection
In this research, various string matching methods were used to determine which is the most effective for creating a string similarity measure for a pathological report. Precisely 12 methods classified according to the working-based method were implemented and are listed as follows:
Edit-based methods: These algorithms determine how many operations are required to change one word into another. In the case of less similarity between two input words, there are more operations to perform [
27]. In this paper, three distances were used (the Hamming distance, Damerau–Levenshtein, and Levenshtein) to test the performance of this type with the pathological reports.
Token-based methods: Instead of whole strings, a collection of tokens is what is expected as the input. Tokens can be single characters, N-Grams, or whole words. Calculating the overlap’s size and normalizing it using a string length measurement is the basis for quantification [
28]. Four distances were used in this paper, the Jaccard distance, N-Gram, bag distance, and Sørensen–Dice coefficient, to test the performance of this type with pathological reports.
Sequence-based methods: The algorithms seek out the most extended sequence in both strings. Therefore, detecting longer similar sequences means a higher similarity score [
28]. In this paper, only the longest common substring algorithm is implemented and compared with other types of algorithms.
Naïve String Matching Algorithm: This compares the provided pattern to every point in the supplied text. The pattern length determines how long each comparison takes to complete, and the number of locations determines how long the text is. In contrast, the modified version of the Naïve algorithm (Rabin–Karp) depends on the hash values for a pattern search to reduce the time [
29]. This paper implemented the Naïve and Rabin–Karp algorithms to find the more suitable one for use in pathological reports analysis.
Fuzzy methods: All of the standard matching algorithms search to find matching with a low threshold of difference or for only exact matching. Therefore, the fuzzy techniques to solve this problem include finding the similarity ratio by the Levenshtein distance [
30]. In this paper, two methods were implemented using the Python language to determine the most effective method for use in the pathological analysis operations.
3.4. Methodology
The proposed system for analyzing pathological reports utilizes a string matching strategy, which is simple and practical due to the limited words found in these reports. Therefore, in this paper, an enhanced algorithm was designed and implemented that can be used in the proposed system that would provide accuracy and fast response. Therefore, in this paper, we design this algorithm based on two essential concepts:
Most of the words found in the CBC tests are of a length not exceeding five letters.
In addition to this, the words usually do not contain special symbols.
Therefore, a coding system was designed based on these cases. In addition, the hashing principle was used depending on the length of the five letters. This saves a lot of time and leads to maintaining accuracy as much as possible. Now, the steps of the proposed system are discussed, step by step, as follows.
First, the reports were obtained from the laboratory as image or pdf files and then were automatically converted into the CSV file format. The reports were then pre-processed by removing the irrelevant text artifacts created by the OCR software and handling the handwriting cases and misspelled error cases that may result from the OCR software or others. The string matching algorithm enables the system to manage errors by matching words depending on the standard data items (stored in the standard dictionary). The data items are a string or number, and these items are used to denote a piece of information within a general patient record (e.g., the patient’s unique ID, age, sex, and other essential information in the CBC report, such as the WBC). Therefore, after implementing 12 different string matching algorithms, the best two algorithms based on implemented results were hybrids with a modified version of the Rabin–Karp algorithm to improve the processing time. However, first, the basic steps of the Rabin–Karp algorithm must be explained, as shown in Algorithm 1, where q’ is a prime number, and
and
are the numbers of characters in the text and the pattern, respectively. The main problems with this algorithm are that its ability to find the exact match patterns depends on the hash values, and it contains some steps that increase the processing time, such as calculating the hash value each time and repeated division and multiplication operations. Moreover, this algorithm requires a large prime integer to prevent hash values that may be identical to different words [
31]. On the other hand, the fuzzy method is used to find the closest words, but it takes much more time than the Rabin algorithm. So, in this paper, a new hybrid algorithm was proposed by combining the Rabin algorithm with the fuzzy method to utilize the advantage and overcome the limitations of these two techniques.
| Algorithm 1 Rabin–Karp Matching Algorithm |
- 1:
Input: T, which is a sequence of terms (words) in which we want to find a pattern, and P, which is the pattern that we want to find it. - 2:
Output: Indexlist, which is a list containing all occurrences of P in T. - 3:
Begin - 4:
Step 1: Define all variables and their initial values. - 5:
Calculate
- 6:
Calculate
- 7:
Calculate
- 8:
Calculate
- 9:
Calculate
- 10:
Calculate
- 11:
Step 2: Find all P in the input T - 12:
for
do - 13:
Calculate - 14:
Calculate - 15:
end for - 16:
for
do - 17:
if then - 18:
if then - 19:
Print (”the pattern found at position:“+ s) - 20:
Indexlist.append(s) - 21:
end if - 22:
end if - 23:
if then - 24:
Calculate - 25:
end if - 26:
end for - 27:
Return Indexlist - 28:
End
|
The main idea is to take advantage of the speed of the Rabin algorithm in processing words with an exact match and specifying the maximum term because the longest word in the standard report is only five letters. So, to design a new equation, as shown in Equation (
1) for generating the hash value, a unique coding system was adopted for report handling to reduce the calculations because these systems deal only with the alphabet (A to Z). Suppose that we want to find the hash value for (term = abcde):
This equation allows us to eliminate the remainder of the division operations, as well as to facilitate the work and reduce the required arithmetic operations. First, calculate the hash value only once for the input term (T). Then, match all of the stored term lengths to the length of T; if the lengths of the terms match, calculate the hash value for the candidate word. Otherwise, move on to the next word. If no exact matches are found, then the fuzzy method should be used to find the closest term and select the one with the smallest different ratio. Therefore, the steps of the proposed Razy algorithm are listed in Algorithm 2.
| Algorithm 2 Razy Algorithm |
- 1:
Input: T, which is a sequence of terms (words) in which we want to find a pattern, and P, which is the pattern that we want to find it. - 2:
Output: Indexlist, which is a list containing all occurrences of P in T. - 3:
Begin - 4:
Step 1: Define all variables and their initial values. - 5:
Set
- 6:
Set
- 7:
Set
- 8:
Set
- 9:
Step 2: Find all positions that give the largest similarity, either exact or approximate. - 10:
Set
- 11:
if
then - 12:
for do - 13:
Calculate - 14:
if then - 15:
Set - 16:
Set - 17:
end if - 18:
end for - 19:
end if - 20:
Return Position - 21:
End main procedure - 22:
procedure Modified-Rabin() - 23:
if
then - 24:
Set - 25:
for do - 26:
Calculate - 27:
if then - 28:
Set - 29:
end if - 30:
end for - 31:
end if - 32:
Return Position - 33:
end procedure - 34:
procedure get_fuzzy_Ratio() - 35:
Calculate
- 36:
Calculate
- 37:
Calculate
- 38:
Return Ratio - 39:
end procedure - 40:
End
|
4. Results
The implemented algorithms are listed in three tables (
Table 1,
Table 2 and
Table 3). The average waiting time (i.e., the amount of time that passes between the moment that a process is requested and finishing it) for each algorithm in these tables is calculated and illustrated in
Table 4, which represents the average execution (implementation) time. Then, depending on the minimum average waiting time values, a new hybrid algorithm is proposed by merging the algorithms to produce a new algorithm to exploit their benefits and overcome their individual limitations.
A comparison was established to measure the execution time efficiency of the proposed algorithm with the optimized Damerau–Levenshtein and dice coefficients using enumeration operations (ODADNEN) [
26], as shown in
Table 5. In addition, to evaluate the proposed algorithm’s efficiency, this paper considers a standard S1 Dataset (only English data) to compare with [
32]. The results of the implementation comparison are shown in
Table 6, which presents the efficiency and flexibility of the proposed method.
On the other hand, the assessment of the performance of the proposed approach is performed in terms of the metrics listed in
Table 7. These metrics include the F1-score, recall, specificity, accuracy, precision, sensitivity, N-value, and P-value [
33,
34,
35,
36,
37]. In these metrics, the true positive, true negative, false positive, and false negative measures used in these metrics are denoted by the
and
symbols, respectively. In this case,
represents the number of words in the dataset that do not match the input term, whereas
represents the number of words that have been compared to the input term and are the same.
The assessment results are presented in
Table 8. In this table, a comparison is performed to show the superiority and effectiveness of the proposed approach. This comparison includes the proposed Razy approach and the Fuzzy approach. The results presented in this table confirm the effectiveness and superiority of the proposed approach. The achieved accuracy using the proposed Razy method is (0.99973894) for the term (The) and (0.999152054) for the term (Good), whereas the corresponding values achieved by the Fuzzy approach are (0.988385718) and (0.9821751). Similarly, the achieved values of the other metrics, such as the sensitivity, specificity, PPV, NPV, and F1-score, are superior for the proposed approach when compared to the currently existing fuzzy method.
To evaluate the statistical significance of the proposed Razy approach, two sets of experiments were conducted. The first set targeted the statistical analysis of the achieved results, and the results of this experiment are presented in
Table 9. In addition, the second set of experiments was conducted using the Wilcoxon signed rank test, and the results are illustrated in
Table 10. As presented in these tables, it can be noted that the proposed approach is stable and has statistical significance based on the recorded results.
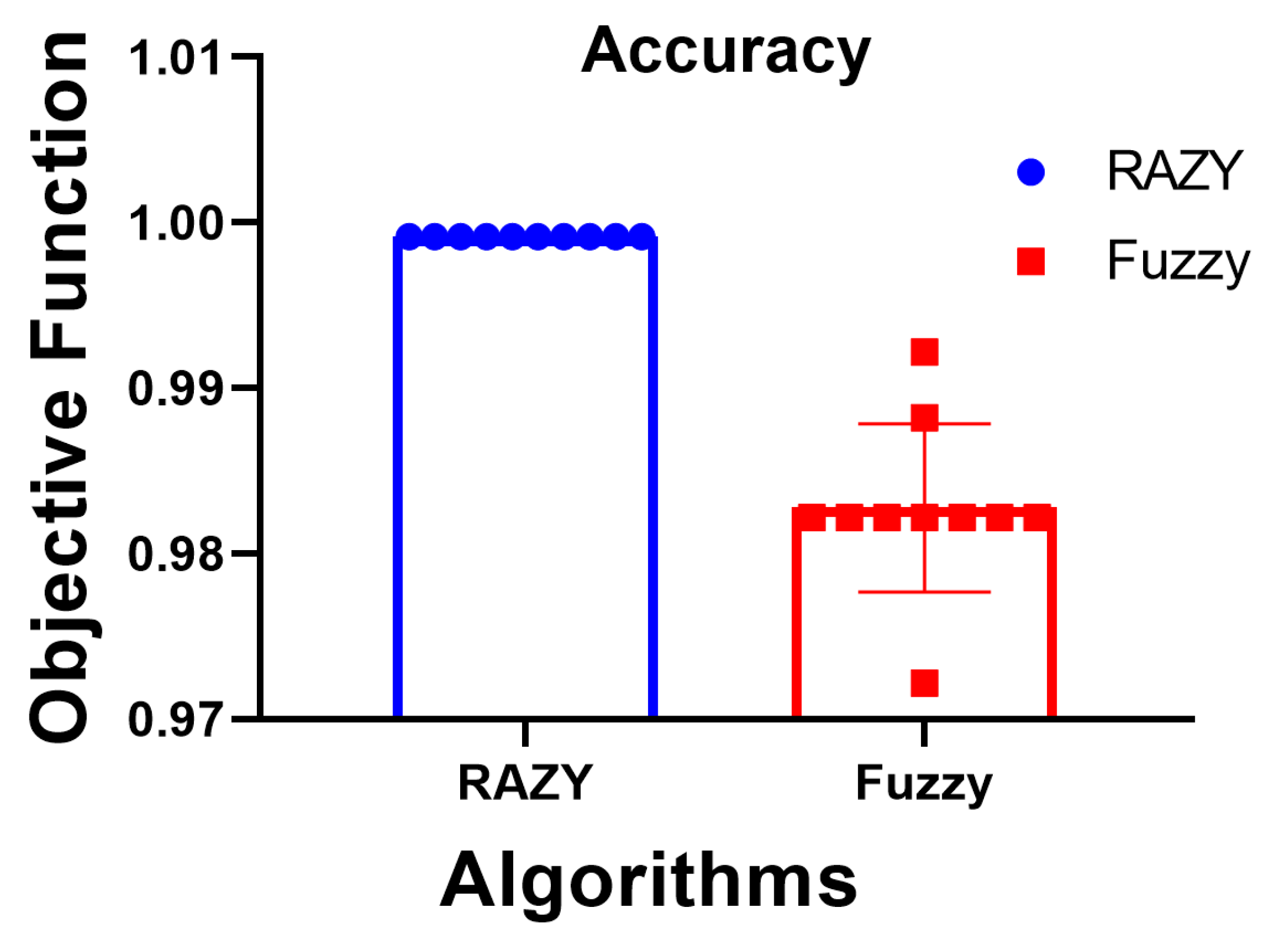
On the other hand, to clearly show the superiority of the proposed approach, the accuracy of the plots for both the proposed Razy and Fuzzy approaches are shown in
Figure 2. As shown in this figure, the accuracy of the proposed approach is stable with a significant value. However, the accuracy of the Razy approach varies from low accuracy to high and is still lower than the proposed approach.
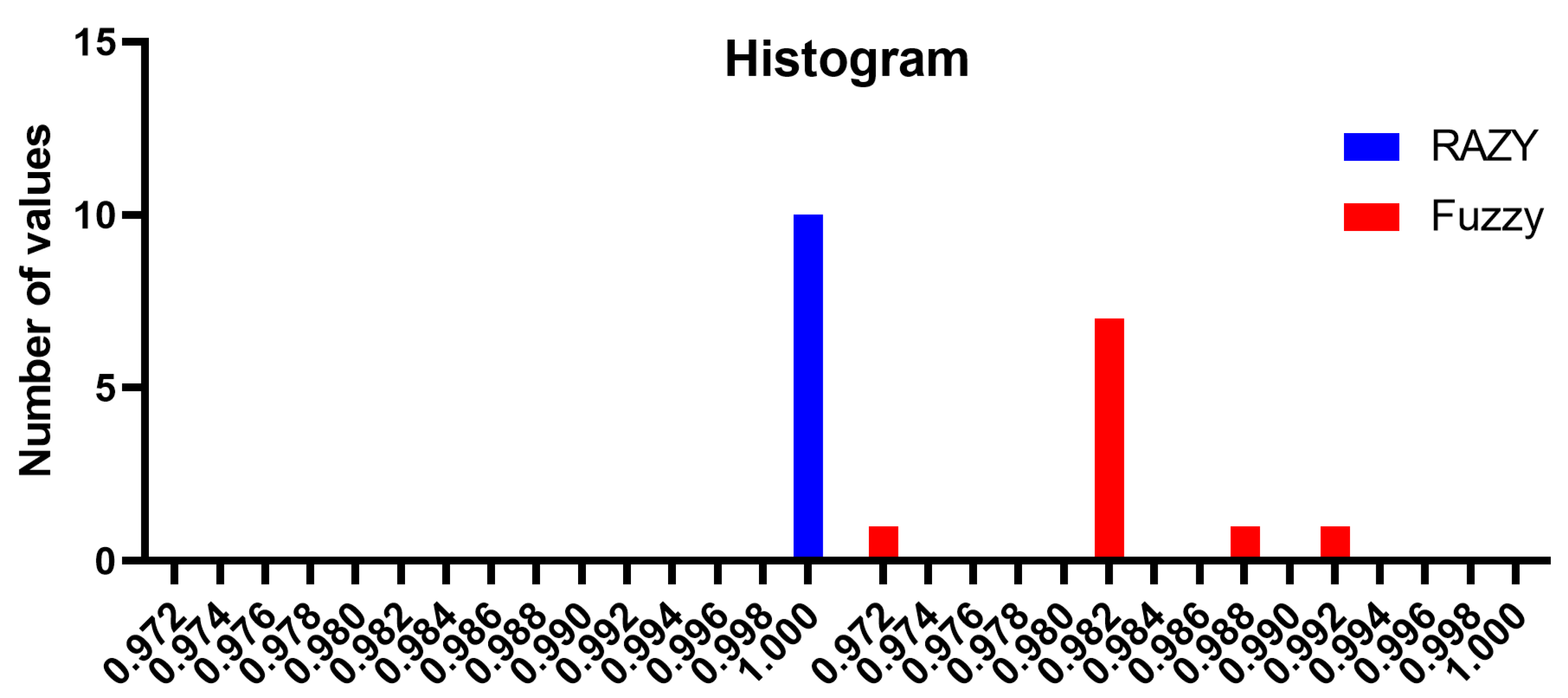
The behavior of the accuracy of the proposed approach in comparison to the Fuzzy approach is clearly obvious in the histogram shown in
Figure 3. In this figure, the histogram of the accuracy using the proposed approach is shown in blue and is almost perfect without variations. However, the accuracy histogram of the Fuzzy approach varies and is lower in value than that of the proposed approach. These results emphasize the superiority and effectiveness of the proposed approach in text mining tasks.
5. Discussion
Text mining involves using string matching algorithms in many subjects, such as document classification (i.e., analysis of their contents and plagiarism detection). However, healthcare analytics systems must optimize large-scale resource administration and availability with the massive increases in resources, users, services, and system contents. Therefore, it might be challenging to identify effective methods for solving the problems and making these decisions in light of the applications and needs. As a result, one of the main goals of this paper is to provide critical analysis of the basis or benchmark methodologies in terms of their advantages and disadvantages. The proposed system can analyze pathological report files (images or pdf) to extract important information to find misspelled words and avoid the information loss caused by approximate string matching or limited by the exact matching approaches to detection of misspelled variations within an acceptable processing time in a large dataset. This paper combines the modified Rabin method with a fuzzy fast similarity search technique capable of using a small different ratio threshold (<1) over the created dictionary of standard terms. The proposed algorithm was implemented using Python, and the findings show that the proposed method can effectively be applied to finding lexically related words in the health sector. The performance of the proposed algorithm was tested using two datasets, and the results are presented in
Table 3 and
Table 4. These tables describe how the proposed method’s efficiency compares to that of its adversaries since it also uses the hash concept with less complex operations to speed up computing and processing. Therefore, the theoretical analysis and experimental results showed that the performance of the proposed (suggested) method was better than the state-of-the-art techniques and is particularly useful for pathological analysis.
The proposed algorithm was designed based on the basic principles available in the words found in pathological reports in terms of the absence of special characters, which affect the encoding system, and the length of the word, which affects the hashing process in the Rabin–Karp algorithm ( i.e., most words are five letters long or less). Moreover, in order to deal with other cases, (words that are longer than five letters; words for which no exact match has been found; whether the file is unclear, contains noise, or contains handwriting words) the fuzzy ratio is used. We can summarize the proposed algorithm as follows:
Razy is the hybrid algorithm that consists of a modified Rabin–Karp algorithm and the fuzzy ratio with a special coding system:
The coding system used for coding symbols depends on the existing symbols in the pathological reports.
A Modified Rabin–Karp Algorithm: Change the process of how to compute the hash values for input strings depending on the number of characters in the coding system and the maximum length for most of the words in the report (five characters).
The ability to find the most similar word using the fuzzy ratio for words with h length maximum and five characters or words that have no match via the Modified Rabin–Karp Algorithm.
However, despite the design and improvement of this algorithm for a specific purpose, which is the analysis of pathological reports, its results were tested on (bible dataset) ordinary English data, and the results are better than the results achieved by the previously published paper, as shown in
Table 6, so we mention that it can be used for other purposes.
6. Conclusions
Doctors track these significant changes by paying attention to blood parameter readings that exceed the normal range. However, slight variations and/or interactions among numerous blood parameters are crucial for identifying abnormal patterns. Therefore, the string similarity can be used to find words (test name) that are similar or identical to words from a standard pathological report in order to collect current patient test values and establish a baseline for the general report. This paper proposed a new algorithm to measure the degree of similarity. This metric, known as Razy string matching, is used to identify two types of matches: exact matches, if any, or approximate matches. Therefore, one of the most important contributions of this research is the development of a matching algorithm for the words found in the CBC tests, which was developed to fit the words in these tests to provide the appropriate time and accuracy for the diagnostic systems for blood diseases. Two additional contributions are building a general report that contains all of the patient’s information (the patient’s medical history) and generating a unique number for each patient through the system, to be included in the report and relied on in dealing with patient data to avoid repetition in the names of patients. The experiments show that an enhanced string similarity measure can be used for both exact and approximate matching with a high accuracy of 99.94% for retrieving results to find the best matching words.