Modeling the Dispersion of E. coli in Waterbodies Due to Urban Sources: A Spatial Approach
Abstract
:1. Introduction
2. Materials and Methods
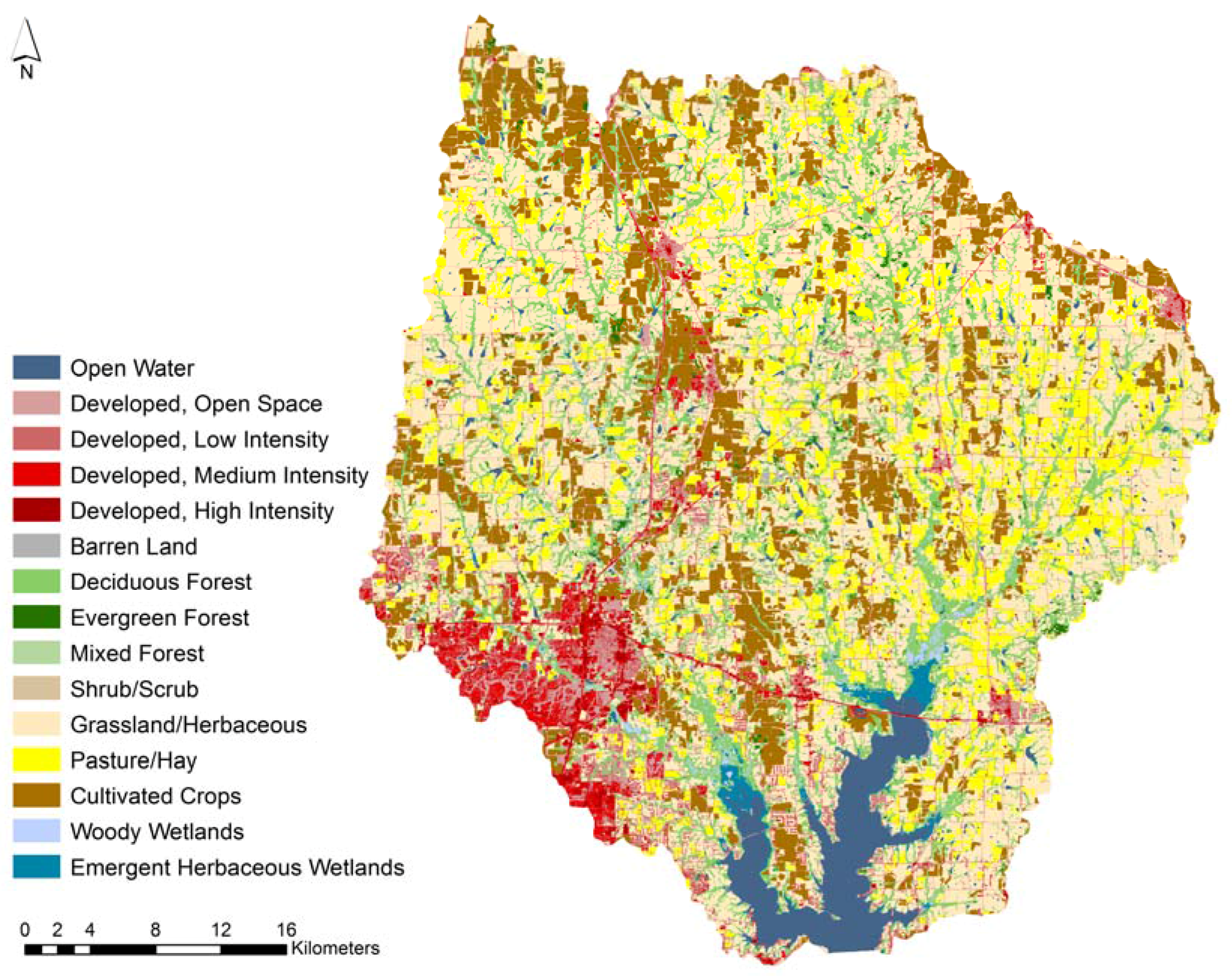
2.1. Spatial Data Layers
2.2. Graphical User Interface
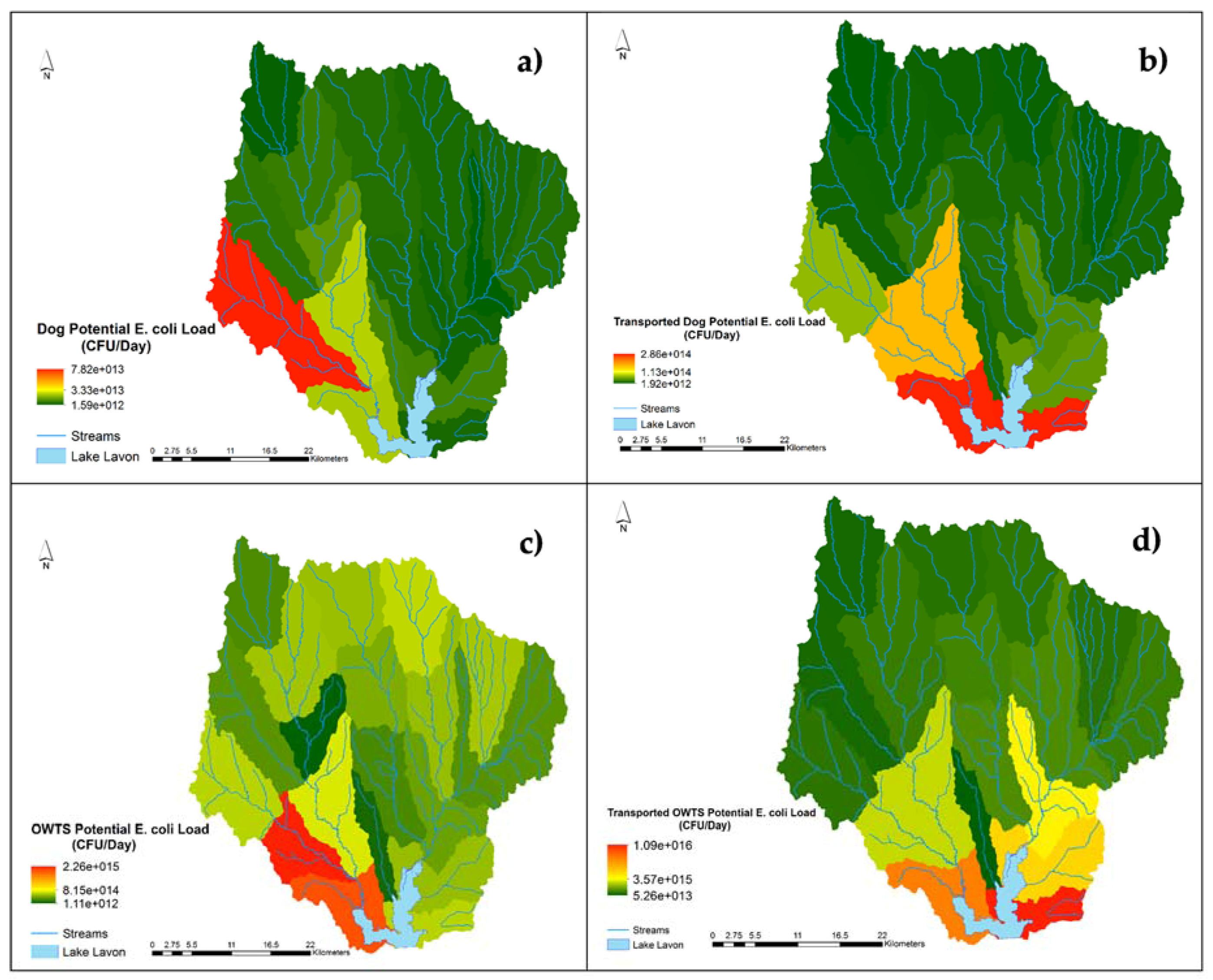
2.3. Potential E. coli Load Module: Dogs
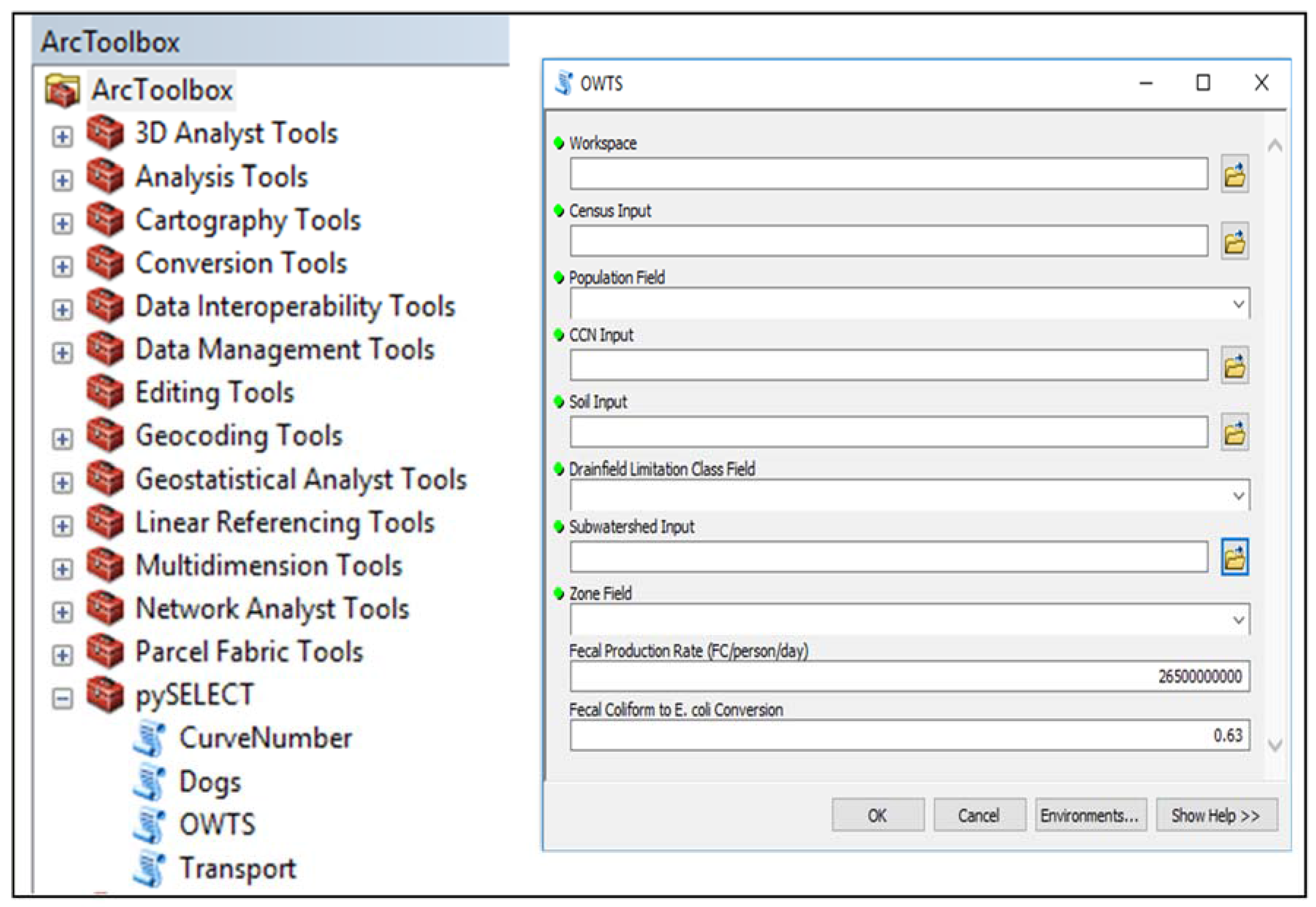
2.4. Potential E. coli Load Module: OWTS
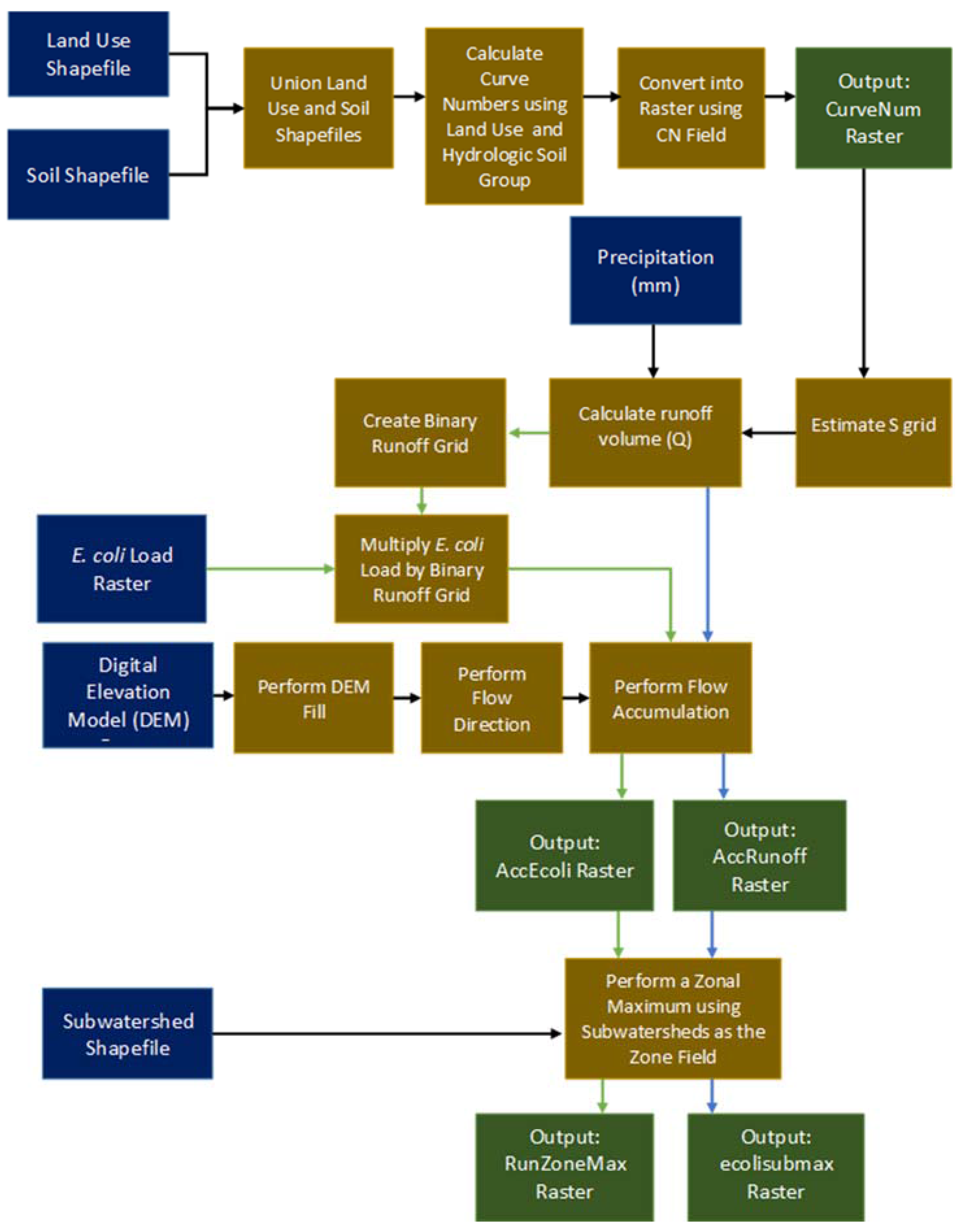
2.5. Rainfall-Runoff and E. coli Transport
3. Tool Verification
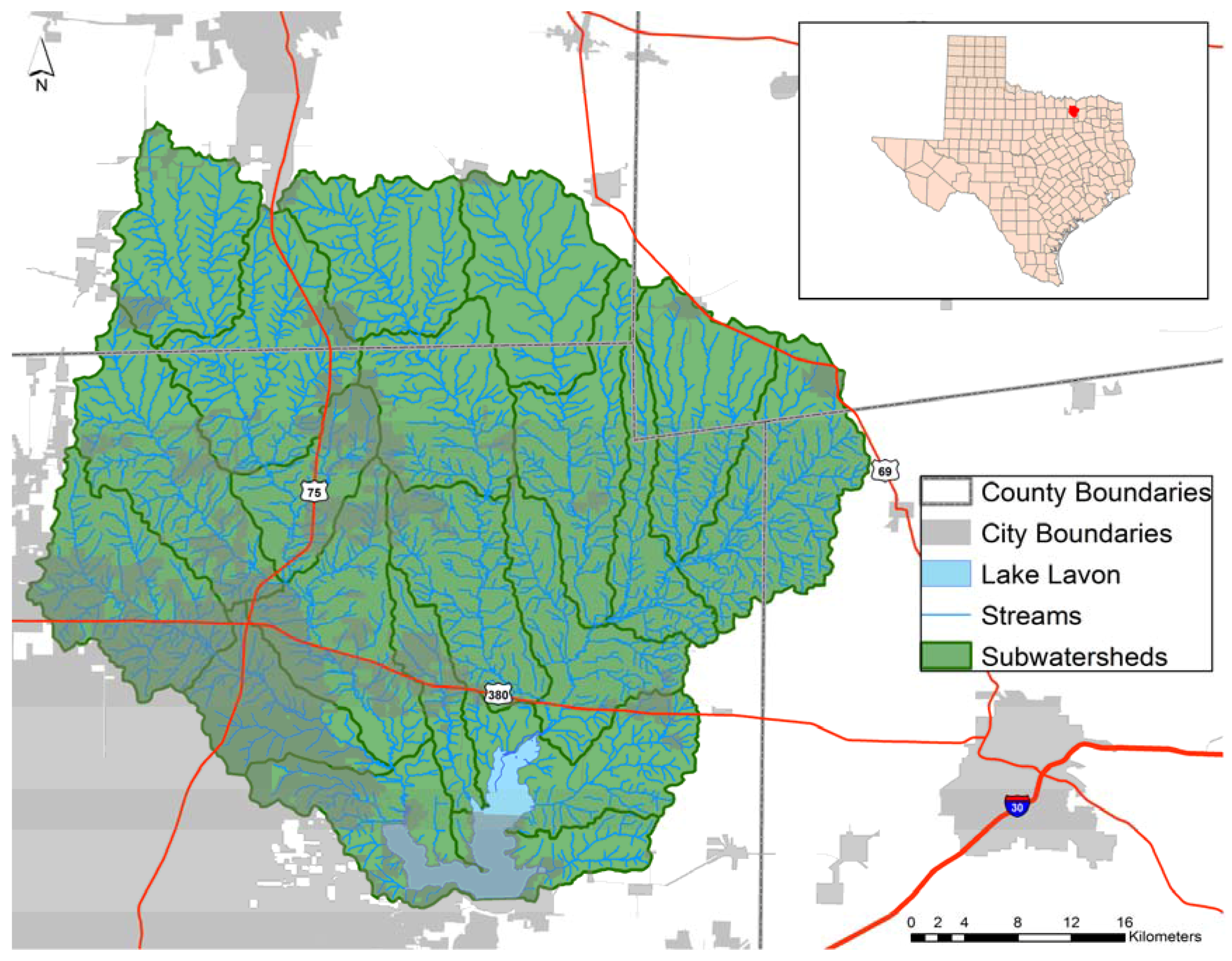
3.1. Study Watershed
3.2. E. coli Transport Model Workflow
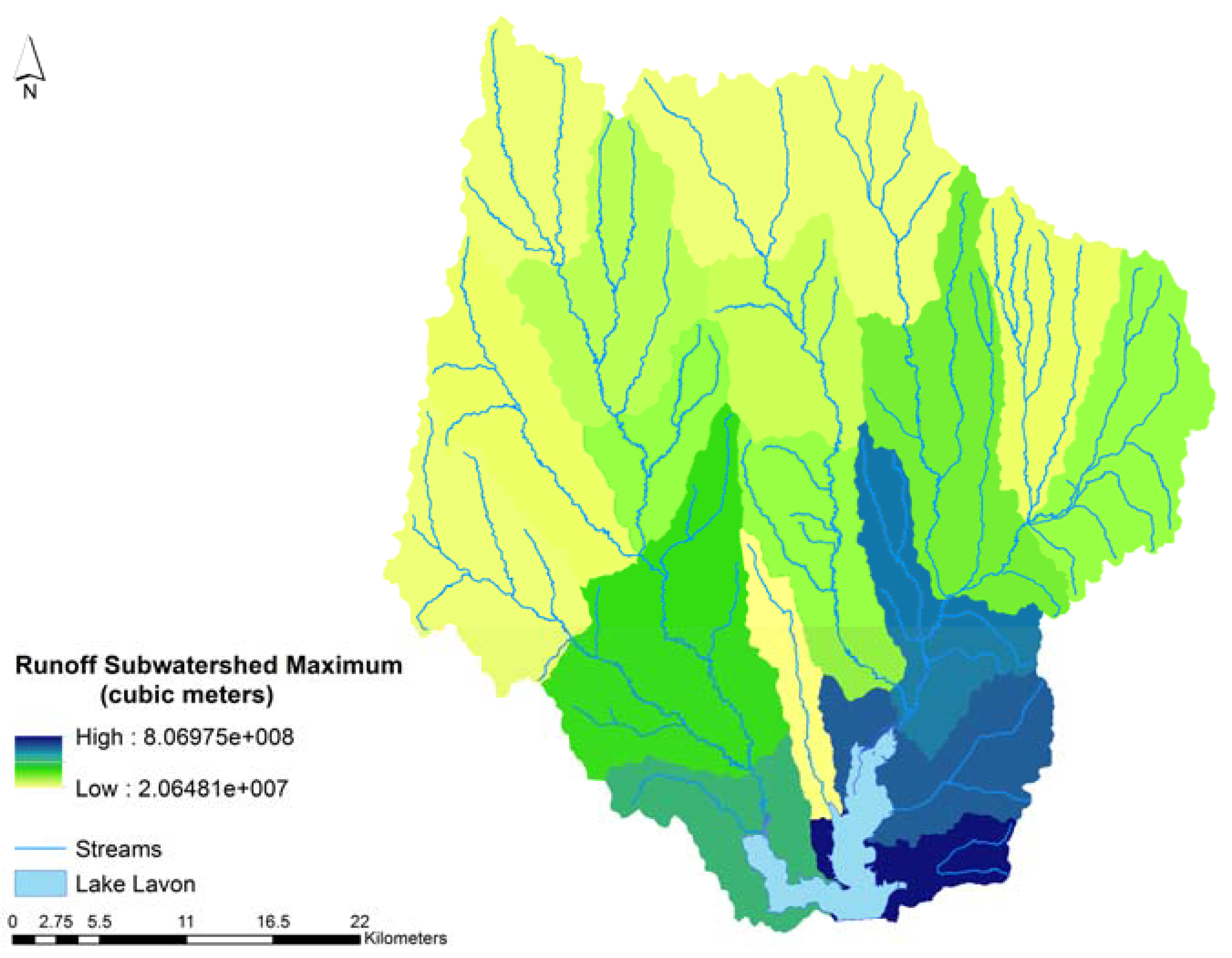
3.3. Results
4. Discussion
4.1. Applications of pySELECT
4.2. Limitations
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- National Summary of State Information. Available online: https://ofmpub.epa.gov/waters10/attains_nation_cy.control (accessed on 31 August 2017).
- Ferguson, C.M.; Croke, B.F.; Beatson, P.J.; Ashbolt, N.J.; Deere, D.A. Development of a process-based model to predict pathogen budgets for the Sydney drinking water catchment. J. Water Health 2007, 5, 187–208. [Google Scholar] [PubMed]
- Frey, S.K.; Topp, E.; Edge, T.; Fall, C.; Gannon, V.; Jokinen, C.; Marti, R.; Neumann, N.; Ruecker, N.; Wilkes, G. Using SWAT, Bacteroidales microbial source tracking markers, and fecal indicator bacteria to predict waterborne pathogen occurrence in an agricultural watershed. Water Res. 2013, 47, 6326–6337. [Google Scholar] [CrossRef] [PubMed]
- Pachepsky, Y.A.; Sadeghi, A.M.; Bradford, S.A.; Shelton, D.R.; Guber, A.K.; Dao, T. Transport and fate of manure-borne pathogens: Modeling perspective. Agric. Water Manag. 2006, 86, 81–92. [Google Scholar] [CrossRef]
- Saleh, A.; Du, B. Evaluation of SWAT and HSPF within BASINS program for the upper North Bosque River watershed in central Texas. Trans. ASAE 2004, 47, 1039–1049. [Google Scholar]
- Teague, A.; Karthikeyan, R.; Babbar-Sebens, M.; Srinivasan, R.; Persyn, R.A. Spatially explicit load enrichment calculation tool to identify potential E. coli sources in watersheds. Trans. ASABE 2009, 52, 1109–1120. [Google Scholar] [CrossRef]
- Riebschleager, K.J.; Karthikeyan, R.; Srinivasan, R.; McKee, K. Estimating Potential Sources in a Watershed Using Spatially Explicit Modeling Techniques. J. Am. Water Res. Assoc. 2012, 48, 745–761. [Google Scholar] [CrossRef]
- Borel, K.E.; Karthikeyan, R.; Smith, P.K.; Srinivasan, R. Predicting E. coli concentrations in surface waters using GIS. J. Nat. Environ. Sci. 2012, 3, 19–33. [Google Scholar]
- Borel, K.E.; Karthikeyan, R.; Smith, P.K.; Gregory, L.F.; Srinivasan, R. Estimating daily potential E. coli loads in rural Texas watersheds using Spatially Explicit Load Enrichment Calculation Tool (SELECT). Tex. Water J. 2012, 3, 42–58. [Google Scholar]
- Borel, K.; Karthikeyan, R.; Berthold, T.A.; Wagner, K. Estimating E. coli and Enterococcus loads in a coastal Texas watershed. Tex. Water J. 2015, 6, 33–44. [Google Scholar]
- Roberts, G.; McFarland, M.; Lloyd, J.; Helton, T.J. Mill Creek Watershed Protection Plan. 2015. Available online: http://millcreek.tamu.edu/files/2014/09/MillCreekWPP.pdf (accessed on 1 September 2017).
- NTMWD; Texas A&M Agrilife Extension Service; TSSWCB Lavon Lake Watershed Protection Plan. 2017. Available online: https://www.ntmwd.com/wp-content/uploads/2017/05/Lavon-Lake-WPP_DRAFT_June2017.pdf (accessed on 1 September 2017).
- Glenn, S.M.; Bare, R.; Neish, B.S. Modeling bacterial load scenarios in a Texas coastal watershed to support decision-making for improving water quality. Tex. Water J. 2017, 8, 57–66. [Google Scholar]
- Coffey, R.; Cummins, E.; Bhreathnach, N.; Flaherty, V.O.; Cormican, M. Development of a pathogen transport model for Irish catchments using SWAT. Agric. Water Manag. 2010, 97, 101–111. [Google Scholar] [CrossRef]
- Jamieson, R.; Gordon, R.; Joy, D.; Lee, H. Assessing microbial pollution of rural surface waters: A review of current watershed scale modeling approaches. Agric. Water Manag. 2004, 70, 1–17. [Google Scholar] [CrossRef]
- Otis, R.; Kreissl, J.F.; Frederick, R.; Goo, R.; Casey, P.; Tonning, B. Onsite Wastewater Treatment Systems Manual (EPA/625/R-00/008); U.S Environmental Protection Agency: Washington, DC, USA, 2002.
- Study to determine the magnitude of, and reasons for, chronically malfunctioning on-site sewage facility systems in Texas. 2001. Available online: https://www.tceq.texas.gov/assets/public/compliance/compliance_support/regulatory/ossf/StudyToDetermine.pdf (accessed on 1 September 2017).
- Mallin, M.A.; Williams, K.E.; Esham, E.C.; Lowe, R.P. Effect of human development on bacteriological water quality in coastal watersheds. Ecol. Appl. 2000, 10, 1047–1056. [Google Scholar] [CrossRef]
- Lin, J.W. Why Python is the next wave in earth sciences computing. Bull. Am. Meteorol. Soc. 2012, 93, 1823–1824. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [PubMed]
- Homer, C.G.; Dewitz, J.A.; Yang, L.; Jin, S.; Danielson, P.; Xian, G.; Coulston, J.; Herold, N.D.; Wickham, J.D.; Megown, K. Completion of the 2011 National Land Cover Database for the conterminous United States-Representing a decade of land cover change information. Photogramm. Eng. Remote Sens. 2015, 81, 345–354. [Google Scholar]
- Soil Data Access. Available online: https://sdmdataaccess.sc.egov.usda.gov (accessed on 31 August 2017).
- TIGER/Line® Shapefiles and TIGER/Line® Files. Available online: https://www.census.gov/geo/maps-data/data/tiger-line.html (accessed on 1 September 2017).
- CCN Mapping Information. Available online: https://www.puc.texas.gov/industry/water/utilities/gis.aspx (accessed on 31 August 2017).
- Geospatial Data Gateway. Available online: http://datagateway.nrcs.usda.gov (accessed on 31 August 2017).
- National Elevation Dataset (NED) 2013. Available online: https://tnris.org/data-catalog/entry/national-elevation-dataset-ned-2013/ (accessed on 31 August 2017).
- Edition, F.; USEPA. Protocol for Developing Pathogen TMDLs; U.S. Environmental Protection Agency: Washington, DC, USA, 2001; p. 132. Available online: https://nepis.epa.gov/Exe/ZyNET.exe/20004QSZ.txt?ZyActionD=ZyDocument&Client=EPA&Index=2000%20Thru%202005&Docs=&Query=&Time=&EndTime=&SearchMethod=1&TocRestrict=n&Toc=&TocEntry=&QField=&QFieldYear=&QFieldMonth=&QFieldDay=&UseQField=&IntQFieldOp=0&ExtQFieldOp=0&XmlQuery=&File=D%3A\ZYFILES\INDEX%20DATA\00THRU05\TXT\00000001\20004QSZ.txt&User=ANONYMOUS&Password=anonymous&SortMethod=h|-&MaximumDocuments=1&FuzzyDegree=0&ImageQuality=r85g16/r85g16/x150y150g16/i500&Display=hpfr&DefSeekPage=x&SearchBack=ZyActionL&Back=ZyActionS&BackDesc=Results%20page&MaximumPages=1&ZyEntry=2 (accessed on 1 September 2017).
- USEPA Recreational water quality criteria. 2012. Available online: https://www.epa.gov/sites/production/files/2015-10/documents/rwqc2012.pdf (accessed on 1 September).
- Soil Survey Manual; Ditzler, C.; Scheffe, K.; Monger, H.C. (Eds.) USDA Handbook 18; Government Printing Office: Washington, DC, USA, 2017.
- USDA-NRCS Urban Hydrology for Small Watersheds. 1986. Available online: ftp://ftp.odot.state.or.us/techserv/Geo-Environmental/Hydraulics/Hydraulics%20Manual/Chapter_07/Chapter_07_appendix_G/Urban_Hydrology_for_Small_Watersheds.pdf (accessed online 1 September 2017).
- Haan, C.T.; Barfield, B.J.; Hayes, J.C. Design Hydrology and Sedimentology for Small Catchments, 3rd ed.; Academic Press: San Diego, CA, USA, 1994. [Google Scholar]
- How Flow Accumulation Works. Available online: https://pro.arcgis.com/en/pro-app/tool-reference/spatial-analyst/how-flow-accumulation-works.htm (accessed on 31 August 2017).
- Lake Lavon Watershed Protection Plan Fact Sheet. Available online: https://www.ntmwd.com/documents/lavon-lake-watershed-protection-plan-fact-sheet (accessed on 31 August 2017).
- TCEQ 2014 Texas Integrated Report; Texas Commission on Environmental Quality: Austin, TX, USA, 2015.
- American Veterinary Medical Association. U.S. pet ownership & demographics sourcebook, 2012 ed.; American Veterinary Medical Association: Schaumburg, IL, USA, 2012; p. 186. [Google Scholar]
- Hershfield, D.M. Rainfall Frequency Atlas of the United States for Durations from 30 Minutes to 24 Hours and Return Periods from 1 to 100 Years; U.S. Department of Commerce: Washington, DC, USA, 1961.

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borel, K.; Swaminathan, V.; Vance, C.; Roberts, G.; Srinivasan, R.; Karthikeyan, R. Modeling the Dispersion of E. coli in Waterbodies Due to Urban Sources: A Spatial Approach. Water 2017, 9, 665. https://doi.org/10.3390/w9090665
Borel K, Swaminathan V, Vance C, Roberts G, Srinivasan R, Karthikeyan R. Modeling the Dispersion of E. coli in Waterbodies Due to Urban Sources: A Spatial Approach. Water. 2017; 9(9):665. https://doi.org/10.3390/w9090665
Chicago/Turabian StyleBorel, Kyna, Vaishali Swaminathan, Cherish Vance, Galen Roberts, Raghavan Srinivasan, and Raghupathy Karthikeyan. 2017. "Modeling the Dispersion of E. coli in Waterbodies Due to Urban Sources: A Spatial Approach" Water 9, no. 9: 665. https://doi.org/10.3390/w9090665






