Tracing COVID-19 Trails in Wastewater: A Systematic Review of SARS-CoV-2 Surveillance with Viral Variants
Abstract
1. Introduction
2. Theoretical Background: The Emergence of SARS-CoV-2 Variants
2.1. Alpha (B.1.1.7 and Q Lineages)
2.2. Delta (B.1.617.2 and AY Lineages)
2.3. Omicron (B.1.1.529 and BA Lineages)
2.4. Other Variants
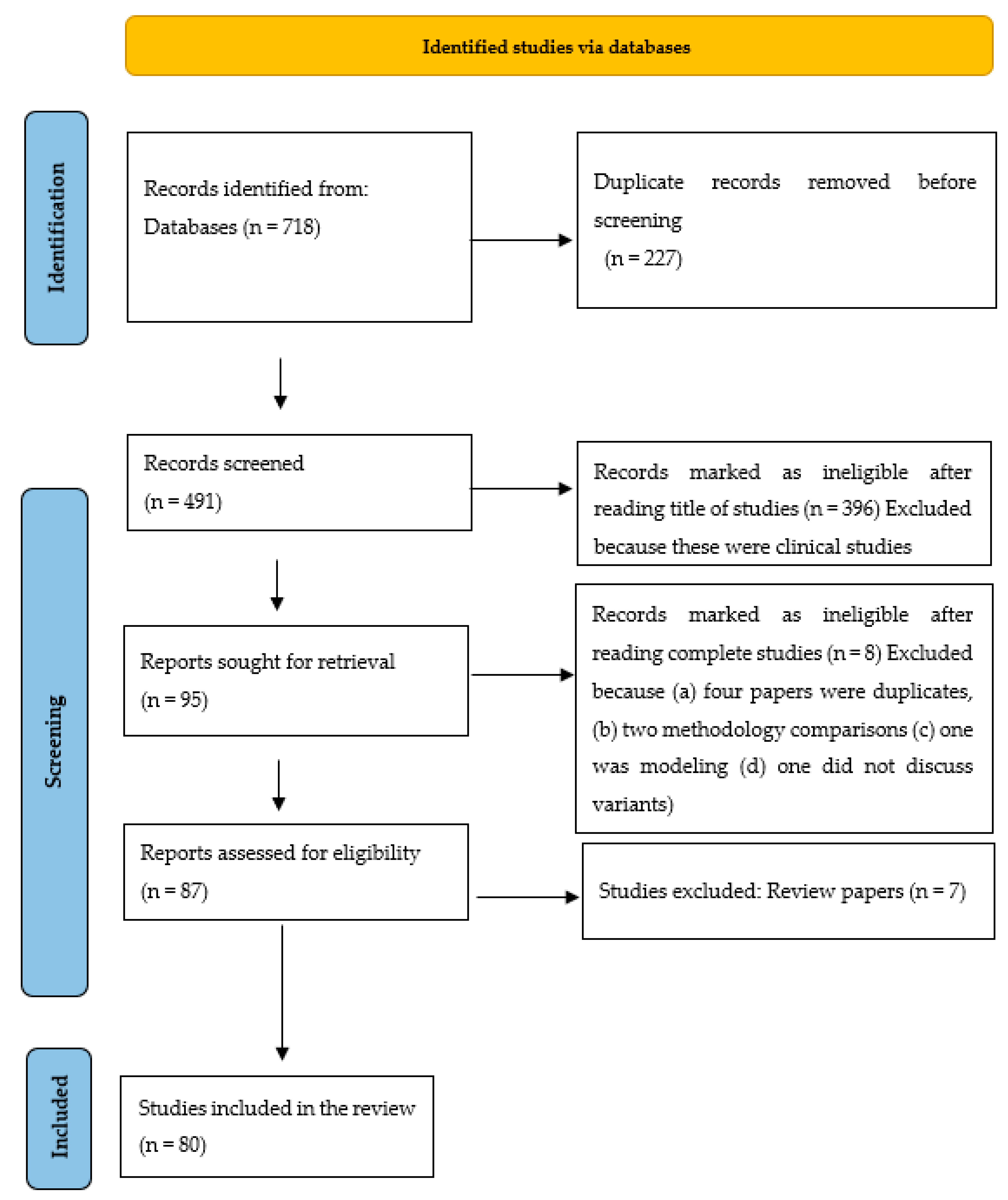
3. Methodology
4. Results
4.1. Geospatial Distribution
4.2. Sampling Techniques
4.3. Concentration Methods
4.4. Analytical Methods to Detect Variants
4.5. Detected Variants
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- CDC. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 27 May 2022).
- Cosar, B.; Karagulleoglu, Z.Y.; Unal, S.; Ince, A.T.; Uncuoglu, D.B.; Tuncer, G.; Kilinc, B.R.; Ozkan, Y.E.; Ozkoc, H.C.; Demir, I.N.; et al. SARS-CoV-2 Mutations and Their Viral Variants. Cytokine Growth Factor Rev. 2022, 63, 10–22. [Google Scholar] [CrossRef]
- Callaway, E. Remember Beta? New Data Reveal Variant’s Deadly Powers. Nature 2021. [Google Scholar] [CrossRef]
- Wilton, T.; Bujaki, E.; Klapsa, D.; Majumdar, M.; Zambon, M.; Fritzsche, M.; Mate, R.; Martin, J. Rapid Increase of SARS-CoV-2 Variant B.1.1.7 Detected in Sewage Samples from England between October 2020 and January 2021. mSystems 2021, 6, e0035321. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Radvak, P.; Kwon, H.J.; Kosikova, M.; Ortega-Rodriguez, U.; Xiang, R.; Phue, J.N.; Shen, R.F.; Rozzelle, J.; Kapoor, N.; Rabara, T.; et al. SARS-CoV-2 B.1.1.7 (Alpha) and B.1.351 (Beta) Variants Induce Pathogenic Patterns in K18-HACE2 Transgenic Mice Distinct from Early Strains. Nat. Commun. 2021, 12, 6559. [Google Scholar] [CrossRef] [PubMed]
- WHO. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 15 December 2022).
- Saguti, F.; Magnil, E.; Enache, L.; Patzi, M.; Johansson, A. Surveillance of Wastewater Revealed Peaks of SARS-CoV-2 Preceding Those of Hospitalized Patients with COVID-19. Water Res. 2020, 189, 116620. [Google Scholar] [CrossRef]
- La Rosa, G.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Iaconelli, M.; Lucentini, L.; Bonadonna, L.; Brusaferro, S.; Brandtner, D.; Fasanella, A.; et al. Rapid Screening for SARS-CoV-2 Variants of Concern in Clinical and Environmental Samples Using Nested RT-PCR Assays Targeting Key Mutations of the Spike Protein. Water Res. 2020, 197, 117104. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Vo, V.; Tillett, R.L.; Papp, K.; Shen, S.; Gu, R.; Gorzalski, A.; Siao, D.; Markland, R.; Chang, C.-L.; Baker, H.; et al. Use of Wastewater Surveillance for Early Detection of Alpha and Epsilon SARS-CoV-2 Variants of Concern and Estimation of Overall COVID-19 Infection Burden. Sci. Total Environ. 2022, 835, 155410. [Google Scholar] [CrossRef]
- Bei, Y.; Pinet, K.; Vrtis, K.B.; Borgaro, J.G.; Sun, L.; Campbell, M.; Apone, L.; Langhorst, B.W.; Nichols, N.M. Overcoming Variant Mutation-Related Impacts on Viral Sequencing and Detection Methodologies. Front. Med. 2022, 9, 989913. [Google Scholar] [CrossRef]
- Ugurel, O.M.; Ata, O.; Turgut-Balik, D. An Updated Analysis of Variations in SARS-CoV-2 Genome. Turk. J. Biol. 2020, 44, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Kwoh, C.K.; Zheng, J. Whole Genome Sequencing Analysis. In Encyclopedia of Bioinformatics and Computational Biology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 176–183. [Google Scholar]
- Tiwari, A.; Lipponen, A.; Hokajärvi, A.-M.; Luomala, O.; Sarekoski, A.; Rytkönen, A.; Österlund, P.; Al-Hello, H.; Juutinen, A.; Miettinen, I.T.; et al. Detection and Quantification of SARS-CoV-2 RNA in Wastewater Influent in Relation to Reported COVID-19 Incidence in Finland. Water Res. 2022, 215, 118220. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First Confirmed Detection of SARS-CoV-2 in Untreated Wastewater in Australia: A Proof of Concept for the Wastewater Surveillance of COVID-19 in the Community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First Detection of SARS-CoV-2 RNA in Wastewater in North America: A Study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cataluña, A.; Chiner-Oms, Á.; Cuevas-Ferrando, E.; Díaz-Reolid, A.; Falcó, I.; Randazzo, W.; Girón-Guzmán, I.; Allende, A.; Bracho, M.A.; Comas, I.; et al. Spatial and Temporal Distribution of SARS-CoV-2 Diversity Circulating in Wastewater. Water Res. 2022, 211, 118007. [Google Scholar] [CrossRef] [PubMed]
- Fontenele, R.S.; Kraberger, S.; Hadfield, J.; Driver, E.M.; Bowes, D.; Holland, L.A.; Faleye, T.O.C.; Adhikari, S.; Kumar, R.; Inchausti, R.; et al. High-Throughput Sequencing of SARS-CoV-2 in Wastewater Provides Insights into Circulating Variants. Water Res. 2021, 205, 117710. [Google Scholar] [CrossRef]
- Islam, M.A.; Rahman, M.A.; Jakariya, M.; Bahadur, N.M.; Hossen, F.; Mukharjee, S.K.; Hossain, M.S.; Tasneem, A.; Haque, M.A.; Sera, F.; et al. A 30-Day Follow-up Study on the Prevalence of SARS-CoV-2 Genetic Markers in Wastewater from the Residence of COVID-19 Patient and Comparison with Clinical Positivity. Sci. Total Environ. 2023, 858, 159350. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Adhikari, S.; Kaya, D.; Islam, M.A.; Malla, B.; Sherchan, S.P.; Al-Mustapha, A.I.; Kumar, M.; Aggarwal, S.; Bhattacharya, P.; et al. Monkeypox Outbreak: Wastewater and Environmental Surveillance Perspective. Sci. Total Environ. 2022, 856, 159166. [Google Scholar] [CrossRef]
- Tiwari, A.; Paakkanen, J.; Österblad, M.; Kirveskari, J.; Hendriksen, R.S.; Heikinheimo, A. Wastewater Surveillance Detected Carbapenemase Enzymes in Clinically Relevant Gram-Negative Bacteria in Helsinki, Finland; 2011–2012. Front. Microbiol. 2022, 13, 887888. [Google Scholar] [CrossRef]
- Martin, J.; Klapsa, D.; Wilton, T.; Zambon, M.; Bentley, E.; Bujaki, E.; Fritzsche, M.; Mate, R.; Majumdar, M. Tracking SARS-CoV-2 in Sewage: Evidence of Changes in Virus Variant Predominance during COVID-19 Pandemic. Viruses 2020, 12, 1144. [Google Scholar] [CrossRef]
- Hokajärvi, A.M.; Rytkönen, A.; Tiwari, A.; Kauppinen, A.; Oikarinen, S.; Lehto, K.M.; Kankaanpää, A.; Gunnar, T.; Al-Hello, H.; Blomqvist, S.; et al. The Detection and Stability of the SARS-CoV-2 RNA Biomarkers in Wastewater Influent in Helsinki, Finland. Sci. Total Environ. 2021, 770, 145274. [Google Scholar] [CrossRef]
- Wurtzer, S.; Waldman, P.; Levert, M.; Cluzel, N.; Almayrac, J.L.; Charpentier, C.; Masnada, S.; Gillon-Ritz, M.; Mouchel, J.M.; Maday, Y.; et al. SARS-CoV-2 Genome Quantification in Wastewaters at Regional and City Scale Allows Precise Monitoring of the Whole Outbreaks Dynamics and Variants Spreading in the Population. Sci. Total Environ. 2022, 810, 152213. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Angel, N.; Bibby, K.; Bivins, A.; Dierens, L.; Edson, J.; Ehret, J.; Gyawali, P.; Hamilton, K.A.; et al. Detection of SARS-CoV-2 RNA in Commercial Passenger Aircraft and Cruise Ship Wastewater: A Surveillance Tool for Assessing the Presence of COVID-19 Infected Travellers. J. Travel Med. 2020, 27, taaa116. [Google Scholar] [CrossRef]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Thompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 Surveillance in Southeastern Virginia Using Wastewater-Based Epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef]
- La Rosa, G.; Brandtner, D.; Mancini, P.; Veneri, C.; Ferraro, G.B.; Bonadonna, L.; Lucentini, L.; Suffredini, E. Key Sars-Cov-2 Mutations of Alpha, Gamma, and Eta Variants Detected in Urban Wastewaters in Italy by Long-Read Amplicon Sequencing Based on Nanopore Technology. Water 2021, 13, 2503. [Google Scholar] [CrossRef]
- Tiwari, A.; Phan, N.; Tandukar, S.; Ashoori, R.; Thakali, O.; Mousazadesh, M.; Dehghani, M.H.; Sherchan, S.P. Persistence and Occurrence of SARS-CoV-2 in Water and Wastewater Environments: A Review of the Current Literature. Environ. Sci. Pollut. Res. 2021, 29, 85658–85668. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in Wastewater: State of the Knowledge and Research Needs. Sci. Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- ECDC. SARS-CoV-2 Variants of Concern as of 8 December 2022. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed on 15 December 2022).
- da Silva, J.C.; Félix, V.B.; Leão, S.A.B.F.; Trindade-Filho, E.M.; Scorza, F.A. New Brazilian Variant of the SARS-CoV-2 (P1/Gamma) of COVID-19 in Alagoas State. Braz. J. Infect. Dis. 2021, 25, 101588. [Google Scholar] [CrossRef] [PubMed]
- Wadman, M. California Coronavirus Strain May Be More Infectious—And Lethal. Science 2021, 23, abh2101. [Google Scholar] [CrossRef]
- Kimura, I.; Kosugi, Y.; Wu, J.; Zahradnik, J.; Yamasoba, D.; Butlertanaka, E.P.; Tanaka, Y.L.; Uriu, K.; Liu, Y.; Morizako, N.; et al. The SARS-CoV-2 Lambda Variant Exhibits Enhanced Infectivity and Immune Resistance. Cell Rep. 2022, 38, 110218. [Google Scholar] [CrossRef]
- Acevedo, M.L.; Gaete-Argel, A.; Alonso-Palomares, L.; de Oca, M.M.; Bustamante, A.; Gaggero, A.; Paredes, F.; Cortes, C.P.; Pantano, S.; Martínez-Valdebenito, C.; et al. Differential Neutralizing Antibody Responses Elicited by CoronaVac and BNT162b2 against SARS-CoV-2 Lambda in Chile. Nat. Microbiol. 2022, 7, 524–529. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, G.I.; Palacios-Pérez, M.; Zamudio, G.S.; Veledíaz, H.F.; Ortega, E.; José, M.V. Neutral Evolution Test of the Spike Protein of SARS-CoV-2 and Its Implications in the Binding to ACE2. Sci. Rep. 2021, 11, 18847. [Google Scholar] [CrossRef] [PubMed]
- Itarte, M.; Bofill-Mas, S.; Martínez-Puchol, S.; Torrell, H.; Ceretó, A.; Carrasco, M.; Forés, E.; Canela, N.; Girones, R.; Rusiñol, M. Looking for a Needle in a Haystack. SARS-CoV-2 Variant Characterization in Sewage. Curr. Opin. Environ. Sci. Health 2021, 24, 100308. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.S.; Vihta, K.-D.; Gethings, O.; Pritchard, E.; Jones, J.; House, T.; Bell, I.; Bell, J.I.; Newton, J.N.; Farrar, J.; et al. Tracking the Emergence of SARS-CoV-2 Alpha Variant in the United Kingdom. N. Engl. J. Med. 2021, 385, 2582–2585. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced Neutralization of SARS-CoV-2 B.1.617 by Vaccine and Convalescent Serum. Cell 2021, 184, 4220–4236.e13. [Google Scholar] [CrossRef]
- Yaniv, K.; Ozer, E.; Lewis, Y.; Kushmaro, A. RT-QPCR Assays for SARS-CoV-2 Variants of Concern in Wastewater Reveals Compromised Vaccination-Induced Immunity. Water Res. 2021, 207, 117808. [Google Scholar] [CrossRef]
- Yaniv, K.; Ozer, E.; Shagan, M.; Lakkakula, S.; Plotkin, N.; Bhandarkar, N.S.; Kushmaro, A. Direct RT-QPCR Assay for SARS-CoV-2 Variants of Concern (Alpha, B.1.1.7 and Beta, B.1.351) Detection and Quantification in Wastewater. Environ. Res. 2021, 201, 111653. [Google Scholar] [CrossRef]
- Hodcroft, E.B.; Zuber, M.; Nadeau, S.; Vaughan, T.G.; Crawford, K.H.D.; Althaus, C.L.; Reichmuth, M.L.; Bowen, J.E.; Walls, A.C.; Corti, D.; et al. Spread of a SARS-CoV-2 Variant through Europe in the Summer of 2020. Nature 2021, 595, 707–712. [Google Scholar] [CrossRef]
- Maida, C.M.; Amodio, E.; Mazzucco, W.; La Rosa, G.; Lucentini, L.; Suffredini, E.; Palermo, M.; Andolina, G.; Iaia, F.R.; Merlo, F.; et al. Wastewater-Based Epidemiology for Early Warning of SARS-CoV-2 Circulation: A Pilot Study Conducted in Sicily, Italy. Int. J. Hyg. Environ. Health 2022, 242, 113948. [Google Scholar] [CrossRef]
- Sharif, S.; Ikram, A.; Khurshid, A.; Salman, M.; Mehmood, N.; Arshad, Y.; Ahmed, J.; Safdar, R.M.; Rehman, L.; Mujtaba, G.; et al. Detection of SARs-CoV-2 in Wastewater Using the Existing Environmental Surveillance Network: A Potential Supplementary System for Monitoring COVID-19 Transmission. PLoS ONE 2021, 16, e0249568. [Google Scholar] [CrossRef]
- Sidik, S.M. Vaccines Protect against Infection from Omicron Subvariant—But Not for Long. Nature 2022. [Google Scholar] [CrossRef]
- Jensen, B.; Luebke, N.; Feldt, T.; Keitel, V.; Brandenburger, T.; Kindgen-Milles, D.; Lutterbeck, M.; Freise, N.F.; Schoeler, D.; Haas, R.; et al. Emergence of the E484K Mutation in SARS-CoV-2-Infected Immunocompromised Patients Treated with Bamlanivimab in Germany. Lancet Reg. Health-Eur. 2021, 8, 100164. [Google Scholar] [CrossRef] [PubMed]
- Kreier, F. Long-COVID Symptoms Less Likely in Vaccinated People, Israeli Data Say. Nature 2022. [Google Scholar] [CrossRef]
- Abdool Karim, S.S.; de Oliveira, T. New SARS-CoV-2 Variants—Clinical, Public Health, and Vaccine Implications. N. Engl. J. Med. 2021, 384, 1866–1868. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef] [PubMed]
- Wurtzer, S.; Levert, M.; Dhenain, E.; Accrombessi, H.; Manco, S.; Fagour, N.; Goulet, M.; Boudaud, N.; Gaillard, L.; Bertrand, I.; et al. From Alpha to Omicron BA.2: New Digital RT-PCR Approach and Challenges for SARS-CoV-2 VOC Monitoring and Normalization of Variant Dynamics in Wastewater. Sci. Total Environ. 2022, 848, 157740. [Google Scholar] [CrossRef]
- Barbé, L.; Schaeffer, J.; Besnard, A.; Jousse, S.; Wurtzer, S.; Moulin, L.; Le Guyader, F.S.; Desdouits, M. SARS-CoV-2 Whole-Genome Sequencing Using Oxford Nanopore Technology for Variant Monitoring in Wastewaters. Front. Microbiol. 2022, 13, 889811. [Google Scholar] [CrossRef]
- Heijnen, L.; Elsinga, G.; de Graaf, M.; Molenkamp, R.; Koopmans, M.P.G.; Medema, G. Droplet Digital RT-PCR to Detect SARS-CoV-2 Signature Mutations of Variants of Concern in Wastewater. Sci. Total Environ. 2021, 799, 149456. [Google Scholar] [CrossRef]
- Ho, J.; Stange, C.; Suhrborg, R.; Wurzbacher, C.; Drewes, J.E.; Tiehm, A. SARS-CoV-2 Wastewater Surveillance in Germany: Long-Term RT-Digital Droplet PCR Monitoring, Suitability of Primer/Probe Combinations and Biomarker Stability. Water Res. 2022, 210, 117977. [Google Scholar] [CrossRef]
- Rubio-Acero, R.; Beyerl, J.; Muenchhoff, M.; Roth, M.S.; Castelletti, N.; Paunovic, I.; Radon, K.; Springer, B.; Nagel, C.; Boehm, B.; et al. Spatially Resolved Qualified Sewage Spot Sampling to Track SARS-CoV-2 Dynamics in Munich—One Year of Experience. Sci. Total Environ. 2021, 797, 149031. [Google Scholar] [CrossRef] [PubMed]
- Schumann, V.-F.; de Castro Cuadrat, R.R.; Wyler, E.; Wurmus, R.; Deter, A.; Quedenau, C.; Dohmen, J.; Faxel, M.; Borodina, T.; Blume, A.; et al. SARS-CoV-2 Infection Dynamics Revealed by Wastewater Sequencing Analysis and Deconvolution. Sci. Total Environ. 2022, 853, 158931. [Google Scholar] [CrossRef]
- Agrawal, S.; Orschler, L.; Tavazzi, S.; Greither, R.; Gawlik, B.M.; Lackner, S. Genome Sequencing of Wastewater Confirms the Arrival of the SARS-CoV-2 Omicron Variant at Frankfurt Airport but Limited Spread in the City of Frankfurt, Germany, in November 2021. Microbiol. Resour. Announc. 2022, 11, e01229-21. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Veneri, C.; Mancini, P.; Bonanno Ferraro, G.; Brandtner, D.; Lucentini, L.; Bonadonna, L.; Rossi, M.; Grigioni, M.; et al. The Rapid Spread of SARS-CoV-2 Omicron Variant in Italy Reflected Early through Wastewater Surveillance. Sci. Total Environ. 2022, 837, 155767. [Google Scholar] [CrossRef]
- Lee, W.L.; Armas, F.; Guarneri, F.; Gu, X.; Formenti, N.; Wu, F.; Chandra, F.; Parisio, G.; Chen, H.; Xiao, A.; et al. Rapid Displacement of SARS-CoV-2 Variant Delta by Omicron Revealed by Allele-Specific PCR in Wastewater. Water Res. 2022, 221, 118809. [Google Scholar] [CrossRef]
- D’Agostino, Y.; Rocco, T.; Ferravante, C.; Porta, A.; Tosco, A.; Cappa, V.M.; Lamberti, J.; Alexandrova, E.; Memoli, D.; Terenzi, I.; et al. Rapid and Sensitive Detection of SARS-CoV-2 Variants in Nasopharyngeal Swabs and Wastewaters. Diagn. Microbiol. Infect. Dis. 2022, 102, 115632. [Google Scholar] [CrossRef]
- Cutrupi, F.; Cadonna, M.; Manara, S.; Postinghel, M.; La Rosa, G.; Suffredini, E.; Foladori, P. The Wave of the SARS-CoV-2 Omicron Variant Resulted in a Rapid Spike and Decline as Highlighted by Municipal Wastewater Surveillance. Environ. Technol. Innov. 2022, 28, 102667. [Google Scholar] [CrossRef] [PubMed]
- Róka, E.; Déri, D.; Khayer, B.; Kis, Z.; Schuler, E.; Magyar, N.; Pályi, B.; Pándics, T.; Vargha, M. SARS-CoV-2 Variant Detection from Wastewater: Rapid Spread of B.1.1.7 Lineage in Hungary. J. Water Health 2022, 20, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Avgeris, M.; Adamopoulos, P.G.; Galani, A.; Xagorari, M.; Gourgiotis, D.; Trougakos, I.P.; Voulgaris, N.; Dimopoulos, M.-A.; Thomaidis, N.S.; Scorilas, A. Novel Nested-Seq Approach for SARS-CoV-2 Real-Time Epidemiology and In-Depth Mutational Profiling in Wastewater. Int. J. Mol. Sci. 2021, 22, 8498. [Google Scholar] [CrossRef]
- Galani, A.; Aalizadeh, R.; Kostakis, M.; Markou, A.; Alygizakis, N.; Lytras, T.; Adamopoulos, P.G.; Peccia, J.; Thompson, D.C.; Kontou, A.; et al. SARS-CoV-2 Wastewater Surveillance Data Can Predict Hospitalizations and ICU Admissions. Sci. Total Environ. 2022, 804, 150151. [Google Scholar] [CrossRef]
- Pechlivanis, N.; Tsagiopoulou, M.; Maniou, M.C.; Togkousidis, A.; Mouchtaropoulou, E.; Chassalevris, T.; Chaintoutis, S.C.; Petala, M.; Kostoglou, M.; Karapantsios, T.; et al. Detecting SARS-CoV-2 Lineages and Mutational Load in Municipal Wastewater and a Use-Case in the Metropolitan Area of Thessaloniki, Greece. Sci. Rep. 2022, 12, 2659. [Google Scholar] [CrossRef]
- Chassalevris, T.; Chaintoutis, S.C.; Koureas, M.; Petala, M.; Moutou, E.; Beta, C.; Kyritsi, M.; Hadjichristodoulou, C.; Kostoglou, M.; Karapantsios, T.; et al. SARS-CoV-2 Wastewater Monitoring Using a Novel PCR-Based Method Rapidly Captured the Delta-to-Omicron ΒA.1 Transition Patterns in the Absence of Conventional Surveillance Evidence. Sci. Total Environ. 2022, 844, 156932. [Google Scholar] [CrossRef]
- Jahn, K.; Dreifuss, D.; Topolsky, I.; Kull, A.; Ganesanandamoorthy, P.; Fernandez-Cassi, X.; Bänziger, C.; Devaux, A.J.; Stachler, E.; Caduff, L.; et al. Early Detection and Surveillance of SARS-CoV-2 Genomic Variants in Wastewater Using COJAC. Nat. Microbiol. 2022, 7, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Caduff, L.; Dreifuss, D.; Schindler, T.; Devaux, A.J.; Ganesanandamoorthy, P.; Kull, A.; Stachler, E.; Fernandez-Cassi, X.; Beerenwinkel, N.; Kohn, T.; et al. Inferring Transmission Fitness Advantage of SARS-CoV-2 Variants of Concern from Wastewater Samples Using Digital PCR, Switzerland, December 2020 through March 2021. Eurosurveillance 2022, 27, 2100806. [Google Scholar] [CrossRef]
- Agrawal, S.; Orschler, L.; Schubert, S.; Zachmann, K.; Heijnen, L.; Tavazzi, S.; Gawlik, B.M.; de Graaf, M.; Medema, G.; Lackner, S. Prevalence and Circulation Patterns of SARS-CoV-2 Variants in European Sewage Mirror Clinical Data of 54 European Cities. Water Res. 2022, 214, 118162. [Google Scholar] [CrossRef]
- Radu, E.; Masseron, A.; Amman, F.; Schedl, A.; Agerer, B.; Endler, L.; Penz, T.; Bock, C.; Bergthaler, A.; Vierheilig, J.; et al. Emergence of SARS-CoV-2 Alpha Lineage and Its Correlation with Quantitative Wastewater-Based Epidemiology Data. Water Res. 2022, 215, 118257. [Google Scholar] [CrossRef] [PubMed]
- Amman, F.; Markt, R.; Endler, L.; Hupfauf, S.; Agerer, B.; Schedl, A.; Richter, L.; Zechmeister, M.; Bicher, M.; Heiler, G.; et al. Viral Variant-Resolved Wastewater Surveillance of SARS-CoV-2 at National Scale. Nat. Biotechnol. 2022, 40, 1814–1822. [Google Scholar] [CrossRef]
- Markt, R.; Endler, L.; Amman, F.; Schedl, A.; Penz, T.; Büchel-Marxer, M.; Grünbacher, D.; Mayr, M.; Peer, E.; Pedrazzini, M.; et al. Detection and Abundance of SARS-CoV-2 in Wastewater in Liechtenstein, and the Estimation of Prevalence and Impact of the B.1.1.7 Variant. J. Water Health 2022, 20, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Herold, M.; D’Hérouël, A.F.; May, P.; Delogu, F.; Wienecke-Baldacchino, A.; Tapp, J.; Walczak, C.; Wilmes, P.; Cauchie, H.-M.; Fournier, G.; et al. Genome Sequencing of SARS-CoV-2 Allows Monitoring of Variants of Concern through Wastewater. Water 2021, 13, 3018. [Google Scholar] [CrossRef]
- Boogaerts, T.; Van den Bogaert, S.; Van Poelvoorde, L.A.E.; El Masri, D.; De Roeck, N.; Roosens, N.H.C.; Lesenfants, M.; Lahousse, L.; Van Hoorde, K.; van Nuijs, A.L.N.; et al. Optimization and Application of a Multiplex Digital PCR Assay for the Detection of SARS-CoV-2 Variants of Concern in Belgian Influent Wastewater. Viruses 2022, 14, 610. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, L.J.; Gonzalez, G.; Sala-Comorera, L.; Martin, N.A.; Byrne, A.; Fennema, S.; Holohan, N.; Kuntamukkula, S.R.; Sarwar, N.; Nolan, T.M.; et al. SARS-CoV-2 Variant Trends in Ireland: Wastewater-Based Epidemiology and Clinical Surveillance. Sci. Total Environ. 2022, 838, 155828. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, L.D.; Richter, S.R.; Midgley, S.E.; Franck, K.T. Detecting SARS-CoV-2 Omicron B.1.1.529 Variant in Wastewater Samples by Using Nanopore Sequencing. Emerg. Infect. Dis. 2022, 28, 1296. [Google Scholar] [CrossRef]
- Mishra, S.; Mindermann, S.; Sharma, M.; Whittaker, C.; Mellan, T.A.; Wilton, T.; Klapsa, D.; Mate, R.; Fritzsche, M.; Zambon, M.; et al. Changing Composition of SARS-CoV-2 Lineages and Rise of Delta Variant in England. EClinicalMedicine 2021, 39, 101064. [Google Scholar] [CrossRef]
- Carcereny, A.; Garcia-Pedemonte, D.; Martínez-Velázquez, A.; Quer, J.; Garcia-Cehic, D.; Gregori, J.; Antón, A.; Andrés, C.; Pumarola, T.; Chacón-Villanueva, C.; et al. Dynamics of SARS-CoV-2 Alpha (B.1.1.7) Variant Spread: The Wastewater Surveillance Approach. Environ. Res. 2022, 208, 112720. [Google Scholar] [CrossRef]
- Novoa, B.; Ríos-Castro, R.; Otero-Muras, I.; Gouveia, S.; Cabo, A.; Saco, A.; Rey-Campos, M.; Pájaro, M.; Fajar, N.; Aranguren, R.; et al. Wastewater and Marine Bioindicators Surveillance to Anticipate COVID-19 Prevalence and to Explore SARS-CoV-2 Diversity by next Generation Sequencing: One-Year Study. Sci. Total Environ. 2022, 833, 155140. [Google Scholar] [CrossRef]
- Rios, G.; Lacoux, C.; Leclercq, V.; Diamant, A.; Lebrigand, K.; Lazuka, A.; Soyeux, E.; Lacroix, S.; Fassy, J.; Couesnon, A.; et al. Monitoring SARS-CoV-2 Variants Alterations in Nice Neighborhoods by Wastewater Nanopore Sequencing. Lancet Reg. Health-Eur. 2021, 10, 100202. [Google Scholar] [CrossRef] [PubMed]
- Wurtz, N.; Revol, O.; Jardot, P.; Giraud-Gatineau, A.; Houhamdi, L.; Soumagnac, C.; Annessi, A.; Lacoste, A.; Colson, P.; Aherfi, S.; et al. Monitoring the Circulation of SARS-CoV-2 Variants by Genomic Analysis of Wastewater in Marseille, South-East France. Pathogens 2021, 10, 1042. [Google Scholar] [CrossRef] [PubMed]
- Rothman, J.A.; Loveless, T.B.; Kapcia, J.; Adams, E.D.; Steele, J.A.; Zimmer-Faust, A.G.; Langlois, K.; Wanless, D.; Griffith, M.; Mao, L.; et al. RNA Viromics of Southern California Wastewater and Detection of SARS-CoV-2 Single-Nucleotide Variants. Appl. Environ. Microbiol. 2021, 87, e01448-21. [Google Scholar] [CrossRef]
- Ai, Y.; Davis, A.; Jones, D.; Lemeshow, S.; Tu, H.; He, F.; Ru, P.; Pan, X.; Bohrerova, Z.; Lee, J. Wastewater SARS-CoV-2 Monitoring as a Community-Level COVID-19 Trend Tracker and Variants in Ohio, United States. Sci. Total Environ. 2021, 801, 149757. [Google Scholar] [CrossRef]
- Wolfe, M.; Hughes, B.; Duong, D.; Chan-Herur, V.; Wigginton, K.R.; White, B.J.; Boehm, A.B. Detection of SARS-CoV-2 Variants Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in Wastewater Settled Solids Using Mutation-Specific Assays Is Associated with Regional Detection of Variants in Clinical Samples. Appl. Environ. Microbiol. 2022, 88, e00045-22. [Google Scholar] [CrossRef]
- Sutton, M.; Radniecki, T.S.; Kaya, D.; Alegre, D.; Geniza, M.; Girard, A.-M.; Carter, K.; Dasenko, M.; Sanders, J.L.; Cieslak, P.R.; et al. Detection of SARS-CoV-2 B.1.351 (Beta) Variant through Wastewater Surveillance before Case Detection in a Community, Oregon, USA. Emerg. Infect. Dis. 2022, 28, 1101. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.T.; Hughes, B.; Wolfe, M.K.; Leon, T.; Duong, D.; Rabe, A.; Kennedy, L.C.; Ravuri, S.; White, B.J.; Wigginton, K.R.; et al. Estimating Relative Abundance of 2 SARS-CoV-2 Variants through Wastewater Surveillance at 2 Large Metropolitan Sites, United States. Emerg. Infect. Dis. 2022, 28, 940–947. [Google Scholar] [CrossRef]
- Oh, C.; Sashittal, P.; Zhou, A.; Wang, L.; El-Kebir, M.; Nguyen, T.H. Design of SARS-CoV-2 Variant-Specific PCR Assays Considering Regional and Temporal Characteristics. Appl. Environ. Microbiol. 2022, 88, e02289-21. [Google Scholar] [CrossRef] [PubMed]
- Brumfield, K.D.; Leddy, M.; Usmani, M.; Cotruvo, J.A.; Tien, C.-T.; Dorsey, S.; Graubics, K.; Fanelli, B.; Zhou, I.; Registe, N.; et al. Microbiome Analysis for Wastewater Surveillance during COVID-19. mBio 2022, 13, e00591-22. [Google Scholar] [CrossRef]
- Layton, B.A.; Kaya, D.; Kelly, C.; Williamson, K.J.; Alegre, D.; Bachhuber, S.M.; Banwarth, P.G.; Bethel, J.W.; Carter, K.; Dalziel, B.D.; et al. Evaluation of a Wastewater-Based Epidemiological Approach to Estimate the Prevalence of SARS-CoV-2 Infections and the Detection of Viral Variants in Disparate Oregon Communities at City and Neighborhood Scales. Environ. Health Perspect. 2022, 130, 067010. [Google Scholar] [CrossRef]
- Silva, C.S.; Tryndyak, V.P.; Camacho, L.; Orloff, M.S.; Porter, A.; Garner, K.; Mullis, L.; Azevedo, M. Temporal Dynamics of SARS-CoV-2 Genome and Detection of Variants of Concern in Wastewater Influent from Two Metropolitan Areas in Arkansas. Sci. Total Environ. 2022, 849, 157546. [Google Scholar] [CrossRef] [PubMed]
- Smyth, D.S.; Trujillo, M.; Gregory, D.A.; Cheung, K.; Gao, A.; Graham, M.; Guan, Y.; Guldenpfennig, C.; Hoxie, I.; Kannoly, S.; et al. Tracking Cryptic SARS-CoV-2 Lineages Detected in NYC Wastewater. Nat. Commun. 2022, 13, 635. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, e02703-20. [Google Scholar] [CrossRef]
- Swift, C.L.; Isanovic, M.; Correa Velez, K.E.; Norman, R.S. Community-Level SARS-CoV-2 Sequence Diversity Revealed by Wastewater Sampling. Sci. Total Environ. 2021, 801, 149691. [Google Scholar] [CrossRef]
- Gregory, D.A.; Wieberg, C.G.; Wenzel, J.; Lin, C.-H.; Johnson, M.C. Monitoring SARS-CoV-2 Populations in Wastewater by Amplicon Sequencing and Using the Novel Program SAM Refiner. Viruses 2021, 13, 1647. [Google Scholar] [CrossRef]
- Ash, K.T.; Alamilla, I.; Li, Y.; Joyner, D.C.; Williams, D.E.; McKay, P.J.; Green, B.M.; Iler, C.; DeBlander, S.E.; Kara-Murdoch, F.; et al. Coding-Complete Genome Sequence of a SARS-CoV-2 Variant Obtained from Raw Sewage at the University of Tennessee—Knoxville Campus. Microbiol. Resour. Announc. 2021, 10, e01049-21. [Google Scholar] [CrossRef]
- Lee, W.L.; Imakaev, M.; Armas, F.; McElroy, K.A.; Gu, X.; Duvallet, C.; Chandra, F.; Chen, H.; Leifels, M.; Mendola, S.; et al. Quantitative SARS-CoV-2 Alpha Variant B.1.1.7 Tracking in Wastewater by Allele-Specific RT-QPCR. Environ. Sci. Technol. Lett. 2021, 8, 675–682. [Google Scholar] [CrossRef]
- Rainey, A.L.; Loeb, J.C.; Robinson, S.E.; Lednicky, J.A.; McPherson, J.; Colson, S.; Allen, M.; Coker, E.S.; Sabo-Attwood, T.; Maurelli, A.T.; et al. Wastewater Surveillance for SARS-CoV-2 in a Small Coastal Community: Effects of Tourism on Viral Presence and Variant Identification among Low Prevalence Populations. Environ. Res. 2022, 208, 112496. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Hughes, B.; Wolfe, M.K.; White, B.J.; Duong, D.; Chan-Herur, V. Regional Replacement of SARS-CoV-2 Variant Omicron BA.1 with BA.2 as Observed through Wastewater Surveillance. Environ. Sci. Technol. Lett. 2022, 9, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Uppal, T.; Hartley, P.; Gorzalski, A.; Pandori, M.; Picker, M.A.; Verma, S.; Pagilla, K. Detecting SARS-CoV-2 variants in wastewater and their correlation with circulating variants in the communities. Sci Rep 2022, 12, 16141. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Glier, M.; Kuchinski, K.; Ross-Van Mierlo, T.; McVea, D.; Tyson, J.R.; Prystajecky, N.; Ziels, R.M. Assessing Multiplex Tiling PCR Sequencing Approaches for Detecting Genomic Variants of SARS-CoV-2 in Municipal Wastewater. mSystems 2021, 6, e01068-21. [Google Scholar] [CrossRef]
- Graber, T.E.; Mercier, É.; Bhatnagar, K.; Fuzzen, M.; D’Aoust, P.M.; Hoang, H.-D.; Tian, X.; Towhid, S.T.; Plaza-Diaz, J.; Eid, W.; et al. Near Real-Time Determination of B.1.1.7 in Proportion to Total SARS-CoV-2 Viral Load in Wastewater Using an Allele-Specific Primer Extension PCR Strategy. Water Res. 2021, 205, 117681. [Google Scholar] [CrossRef]
- Corchis-Scott, R.; Geng, Q.; Seth, R.; Ray, R.; Beg, M.; Biswas, N.; Charron, L.; Drouillard, K.D.; D’Souza, R.; Heath, D.D.; et al. Averting an Outbreak of SARS-CoV-2 in a University Residence Hall through Wastewater Surveillance. Microbiol. Spectr. 2021, 9, e00792-21. [Google Scholar] [CrossRef]
- Hubert, C.R.J.; Acosta, N.; Waddell, B.J.M.; Hasing, M.E.; Qiu, Y.; Fuzzen, M.; Harper, N.B.J.; Bautista, M.A.; Gao, T.; Papparis, C.; et al. Tracking Emergence and Spread of SARS-CoV-2 Omicron Variant in Large and Small Communities by Wastewater Monitoring in Alberta, Canada. Emerg. Infect. Dis. 2022, 28, 1770–1776. [Google Scholar] [CrossRef] [PubMed]
- Lawal, O.U.; Zhang, L.; Parreira, V.R.; Brown, R.S.; Chettleburgh, C.; Dannah, N.; Delatolla, R.; Gilbride, K.A.; Graber, T.E.; Islam, G.; et al. Metagenomics of Wastewater Influent from Wastewater Treatment Facilities across Ontario in the Era of Emerging SARS-CoV-2 Variants of Concern. Microbiol. Resour. Announc. 2022, 11, e00362-22. [Google Scholar] [CrossRef]
- Oloye, F.F.; Xie, Y.; Asadi, M.; Cantin, J.; Challis, J.K.; Brinkmann, M.; McPhedran, K.N.; Kristian, K.; Keller, M.; Sadowski, M.; et al. Rapid Transition between SARS-CoV-2 Variants of Concern Delta and Omicron Detected by Monitoring Municipal Wastewater from Three Canadian Cities. Sci. Total Environ. 2022, 841, 156741. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Sun, J.; Yang, M.I.; Gibson, R.M.; Arts, E.J.; Olabode, A.S.; Poon, A.F.Y.; Wang, X.; Wheeler, A.R.; Edwards, E.A.; et al. Early Warning Measurement of SARS-CoV-2 Variants of Concern in Wastewaters by Mass Spectrometry. Environ. Sci. Technol. Lett. 2022, 9, 638–644. [Google Scholar] [CrossRef]
- Peterson, S.W.; Lidder, R.; Daigle, J.; Wonitowy, Q.; Dueck, C.; Nagasawa, A.; Mulvey, M.R.; Mangat, C.S. RT-QPCR Detection of SARS-CoV-2 Mutations S 69–70 Del, S N501Y and N D3L Associated with Variants of Concern in Canadian Wastewater Samples. Sci. Total Environ. 2022, 810, 151283. [Google Scholar] [CrossRef] [PubMed]
- Napit, R.; Manandhar, P.; Chaudhary, A.; Shrestha, B.; Poudel, A.; Raut, R.; Pradhan, S.; Raut, S.; Mathema, S.; Rajbhandari, R.; et al. Rapid Genomic Surveillance of SARS-CoV-2 in a Dense Urban Community Using Environmental (Sewage) Samples. medRxiv 2021. [Google Scholar] [CrossRef]
- Otero, M.C.B.; Murao, L.A.E.; Limen, M.A.G.; Gaite, P.L.A.; Bacus, M.G.; Acaso, J.T.; Corazo, K.; Knot, I.E.; Sajonia, H.; de los Reyes, F.L.; et al. Wastewater-Based Epidemiology and Whole-Genome Sequencing for Community-Level Surveillance of SARS-CoV-2 in Selected Urban Communities of Davao City, Philippines: A Pilot Study. medRxiv 2021. [Google Scholar] [CrossRef]
- Malla, B.; Thakali, O.; Shrestha, S.; Segawa, T.; Kitajima, M.; Haramoto, E. Application of a High-Throughput Quantitative PCR System for Simultaneous Monitoring of SARS-CoV-2 Variants and Other Pathogenic Viruses in Wastewater. Sci. Total Environ. 2022, 853, 158659. [Google Scholar] [CrossRef]
- Erster, O.; Mendelson, E.; Kabat, A.; Levy, V.; Mannasse, B.; Assraf, H.; Azar, R.; Ali, Y.; Bucris, E.; Bar-Ilan, D.; et al. Specific Detection of SARS-CoV-2 Variants B.1.1.7 (Alpha) and B.1.617.2 (Delta) Using a One-Step Quantitative PCR Assay. Microbiol. Spectr. 2022, 10, e02176-21. [Google Scholar] [CrossRef]
- Bar-Or, I.; Indenbaum, V.; Weil, M.; Elul, M.; Levi, N.; Aguvaev, I.; Cohen, Z.; Levy, V.; Azar, R.; Mannasse, B.; et al. National Scale Real-Time Surveillance of SARS-CoV-2 Variants Dynamics by Wastewater Monitoring in Israel. Viruses 2022, 14, 1229. [Google Scholar] [CrossRef]
- Joshi, M.; Kumar, M.; Srivastava, V.; Kumar, D.; Rathore, D.S.; Pandit, R.; Graham, D.W.; Joshi, C.G. Genetic Sequencing Detected the SARS-CoV-2 Delta Variant in Wastewater a Month Prior to the First COVID-19 Case in Ahmedabad (India). Environ. Pollut. 2022, 310, 119757. [Google Scholar] [CrossRef]
- Nag, A.; Arora, S.; Sinha, V.; Meena, E.; Sutaria, D.; Gupta, A.B.; Medicherla, K.M. Monitoring of SARS-CoV-2 Variants by Wastewater-Based Surveillance as a Sustainable and Pragmatic Approach—A Case Study of Jaipur (India). Water 2022, 14, 297. [Google Scholar] [CrossRef]
- Dharmadhikari, T.; Rajput, V.; Yadav, R.; Boargaonkar, R.; Patil, D.; Kale, S.; Kamble, S.P.; Dastager, S.G.; Dharne, M.S. High Throughput Sequencing Based Direct Detection of SARS-CoV-2 Fragments in Wastewater of Pune, West India. Sci. Total Environ. 2022, 807, 151038. [Google Scholar] [CrossRef]
- Barbosa, M.R.F.; Garcia, S.C.; de Castro Bruni, A.; Machado, F.S.; de Oliveira, R.X.; Dropa, M.; da Costa, A.C.; Leal, E.; Brandão, C.J.; da Silva, R.L.O.; et al. One-Year Surveillance of SARS-CoV-2 in Wastewater from Vulnerable Urban Communities in Metropolitan São Paulo, Brazil. J. Water Health 2022, 20, 471–490. [Google Scholar] [CrossRef]
- Masachessi, G.; Castro, G.; Cachi, A.M.; de los Ángeles Marinzalda, M.; Liendo, M.; Pisano, M.B.; Sicilia, P.; Ibarra, G.; Rojas, R.M.; López, L.; et al. Wastewater Based Epidemiology as a Silent Sentinel of the Trend of SARS-CoV-2 Circulation in the Community in Central Argentina. Water Res. 2022, 219, 118541. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Sharma, J.R.; Ramharack, P.; Mangwana, N.; Kinnear, C.; Viraragavan, A.; Glanzmann, B.; Louw, J.; Abdelatif, N.; Reddy, T.; et al. Tracking the Circulating SARS-CoV-2 Variant of Concern in South Africa Using Wastewater-Based Epidemiology. Sci. Rep. 2022, 12, 1182. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Smith, W.J.M.; Metcalfe, S.; Stephens, M.; Jennison, A.V.; Moore, F.A.J.; Bourke, J.; Schlebusch, S.; McMahon, J.; et al. Detection of the Omicron (B.1.1.529) Variant of SARS-CoV-2 in Aircraft Wastewater. Sci. Total Environ. 2022, 820, 153171. [Google Scholar] [CrossRef] [PubMed]
- Bertels, X.; Demeyer, P.; Van den Bogaert, S.; Boogaerts, T.; van Nuijs, A.L.N.; Delputte, P.; Lahousse, L. Factors Influencing SARS-CoV-2 RNA Concentrations in Wastewater up to the Sampling Stage: A Systematic Review. Sci. Total Environ. 2022, 820, 153290. [Google Scholar] [CrossRef]
- Kmush, B.L.; Monk, D.; Green, H.; Sachs, D.A.; Zeng, T.; Larsen, D.A. Comparability of 24-Hour Composite and Grab Samples for Detection of SARS-2-CoV RNA in Wastewater. FEMS Microbes 2022, 3, xtac017. [Google Scholar] [CrossRef]
- Barril, P.A.; Pianciola, L.A.; Mazzeo, M.; Ousset, M.J.; Jaureguiberry, M.V.; Alessandrello, M.; Sánchez, G.; Oteiza, J.M. Evaluation of Viral Concentration Methods for SARS-CoV-2 Recovery from Wastewaters. Sci. Total Environ. 2021, 756, 144105. [Google Scholar] [CrossRef]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef]
- Tiwari, A.; Ahmed, W.; Oikarinen, S.; Sherchan, S.P.; Heikinheimo, A.; Jiang, G.; Simpson, S.L.; Greaves, J.; Bivins, A. Application of Digital PCR for Public Health-Related Water Quality Monitoring. Sci. Total Environ. 2022, 837, 155663. [Google Scholar] [CrossRef]
- Mazumder, P.; Dash, S.; Honda, R.; Sonne, C.; Kumar, M. Sewage Surveillance for SARS-CoV-2: Molecular Detection, Quantification, and Normalization Factors. Curr. Opin. Environ. Sci. Health 2022, 28, 100363. [Google Scholar] [CrossRef]
- ECDC; WHO. Methods for the Detection and Characterisation of SARS-CoV-2 Variants–Second Update; 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/methods-detection-and-characterisation-sars-cov-2-variants-second-update (accessed on 7 December 2022).
- Ahmed, W.; Smith, W.J.M.; Metcalfe, S.; Jackson, G.; Choi, P.M.; Morrison, M.; Field, D.; Gyawali, P.; Bivins, A.; Bibby, K.; et al. Comparison of RT-QPCR and RT-DPCR Platforms for the Trace Detection of SARS-CoV-2 RNA in Wastewater. ACS ES&T Water 2022, 2, 1871–1880. [Google Scholar] [CrossRef]
- GISAID. Available online: https://gisaid.org/. (accessed on 7 December 2022).
- Thakali, O.; Raya, S.; Malla, B.; Tandukar, S.; Tiwari, A.; Sherchan, S.P.; Sherchand, J.B.; Haramoto, E. Pilot Study on Wastewater Surveillance of Dengue Virus RNA: Lessons, Challenges, and Implications for Future Research. Environ. Chall. 2022, 9, 100614. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Stephens, M.; Metcalfe, S.; Smith, W.J.M.; Sirikanchana, K.; Kitajima, M.; Simpson, S.L. Occurrence of Multiple Respiratory Viruses in Wastewater in Queensland, Australia: Potential for Community Disease Surveillance. Sci. Total Environ. 2023, 864, 161023. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Kurittu, P.; Al-Mustapha, A.I.; Heljanko, V.; Johansson, V.; Thakali, O.; Mishra, S.K.; Lehto, K.-M.; Lipponen, A.; Oikarinen, S.; et al. Wastewater Surveillance of Antibiotic-Resistant Bacterial Pathogens: A Systematic Review. Front. Microbiol. 2022, 13, 977106. [Google Scholar] [CrossRef]
- Adhikari, S.; Halden, R.U. Opportunities and Limits of Wastewater-Based Epidemiology for Tracking Global Health and Attainment of UN Sustainable Development Goals. Environ. Int. 2022, 163, 107217. [Google Scholar] [CrossRef]
- Wolfe, M.K.; Duong, D.; Bakker, K.M.; Ammerman, M.; Mortenson, L.; Hughes, B.; Arts, P.; Lauring, A.S.; Fitzsimmons, W.J.; Bendall, E.; et al. Wastewater-Based Detection of Two Influenza Outbreaks. Environ. Sci. Technol. Lett. 2022, 9, 687–692. [Google Scholar] [CrossRef]



| WHO Label/Pango Lineage | Country First Detected | Spike Mutations of Interest | Outbreak Countries | Major Outbreak Peaks | Classification (WHO) during November 2022 | Outbreak Condition during November 2022 |
|---|---|---|---|---|---|---|
| Alpha/B.1.1.7 | United Kingdom, September 2020 | N501Y, D614G, P681H | At least 189 countries, predominant in the US, India, Sweden, France, Spain, Australia, Nigeria, and so on (1.2 million cases globally reported). | November 2020 to August 2021 | VOC: 29 December 2020 VBM: 21 September 2021 PVOC: 9 March 2022 | Drastically reduced circulation globally, with almost no reporting at the time of writing the manuscript. |
| Delta/B.1.617.2 | India, October 2020 | L452R, T478K, D614G, P681R | At least 208 countries, predominant in the US, UK, Japan, Italy, India, Germany, Canada, Denmark, France, and so on (4.4 million cases globally). | May 2021–January 2022 | VOC: 15 June 2021 VBM: 14 April 2022, PVOC: 7 June 2022 | Abundance is very low at the time of writing the manuscript. |
| Beta/B.1.351 | South Africa, May 2020 | K417N, E484K, N501Y, D614G, A701V | At least 127 countries, predominant in South Africa, the US, India, Sweden, France, Spain, Australia, Nigeria, Iran, and so on (43,000 cases globally). | November 2020 to August 2021 | VOC: 29 December 2020 VBM: 21 September 2021, PVOC: 9 March 2022 | Drastically reduced circulation globally, with almost no reporting at the time of writing the manuscript. |
| Gamma/P.1 | Brazil, November 2020 | K417T, E484K, N501Y, D614G, H655Y | At least 93 countries, predominant in the US, Canada, Brazil, Argentina, Chile, Italy, Peru, Mexico, Sweden, South Korea, Venezuela, and so on (74,300 cases globally). | Feb 2021–November 2021 | VOC: 29 December 2020 VBM: 21 September 2021 PVOC: 9 March 2022 | No longer detected or detected at extremely low levels globally. |
| Epsilon/B.1.427, B.1429 | California USA, July 2020 | I4205V and D1183Y in the ORF1ab gene, and S13I, W152C, L452R in the spike protein’s S-gene | At least 45 countries. | November 2020–March 2021 | VOC, March 2021. VBM-September 2021. Previously circulating VOI: March 2022 (WHO), | After an initial increase, its prevalence rapidly decreased from February 2021 and was outcompeted by the more transmissible Alpha variant. |
| Lambda/C.37 | Peru, August 2020 | Virus’s spike protein code: G75V, T76I, Δ246–252, L452Q, F490S, D614G and T859N | At least 45 countries (predominant in Peru, Chile, US). Total global cases of less than 10,000. | November 2020–November 2021 | VOI June 2021 | No longer reported. |
| Omicron/BA.2, BA.4, BA.5, BA.2.75, BQ.1, XBB | South Africa and Botswana November, 2021 | BA.2 (y*), BA.4 (L452R, F486V, R493Q), BA.5 (L452R, F486V, R493Q). BA.2.75 (z**), BQ.1 (K444T, N460K), XBB (N460K, F490S) | At least 208 countries, predominant in the US, UK, Denmark, Canada, India, Japan, Germany, France, and so on (6.2 million cases globally, 14 December 2022). | November 2021-Currently circulating lineages: BA.2, BA.4, BA.5, and their recombinants and sub-lineages | BA.2, BA.4, and BA.5 are VOC, and BA.2.75, BQ.1, XBB are current VOI at the time of drafting the manuscript, December 2022. | Many lineages are currently going around the world (83,046 in the last four weeks of 14 December 2022). |
| Sequencing | RT-qPCR/RT-dPCR | |
|---|---|---|
| Pros |
|
|
| Cons |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, A.; Adhikari, S.; Zhang, S.; Solomon, T.B.; Lipponen, A.; Islam, M.A.; Thakali, O.; Sangkham, S.; Shaheen, M.N.F.; Jiang, G.; et al. Tracing COVID-19 Trails in Wastewater: A Systematic Review of SARS-CoV-2 Surveillance with Viral Variants. Water 2023, 15, 1018. https://doi.org/10.3390/w15061018
Tiwari A, Adhikari S, Zhang S, Solomon TB, Lipponen A, Islam MA, Thakali O, Sangkham S, Shaheen MNF, Jiang G, et al. Tracing COVID-19 Trails in Wastewater: A Systematic Review of SARS-CoV-2 Surveillance with Viral Variants. Water. 2023; 15(6):1018. https://doi.org/10.3390/w15061018
Chicago/Turabian StyleTiwari, Ananda, Sangeet Adhikari, Shuxin Zhang, Tamunobelema B. Solomon, Anssi Lipponen, Md. Aminul Islam, Ocean Thakali, Sarawut Sangkham, Mohamed N. F. Shaheen, Guangming Jiang, and et al. 2023. "Tracing COVID-19 Trails in Wastewater: A Systematic Review of SARS-CoV-2 Surveillance with Viral Variants" Water 15, no. 6: 1018. https://doi.org/10.3390/w15061018
APA StyleTiwari, A., Adhikari, S., Zhang, S., Solomon, T. B., Lipponen, A., Islam, M. A., Thakali, O., Sangkham, S., Shaheen, M. N. F., Jiang, G., Haramoto, E., Mazumder, P., Malla, B., Kumar, M., Pitkänen, T., & Sherchan, S. P. (2023). Tracing COVID-19 Trails in Wastewater: A Systematic Review of SARS-CoV-2 Surveillance with Viral Variants. Water, 15(6), 1018. https://doi.org/10.3390/w15061018











