SARS-CoV-2 Wastewater Surveillance in Ten Cities from Mexico
Abstract
:1. Introduction
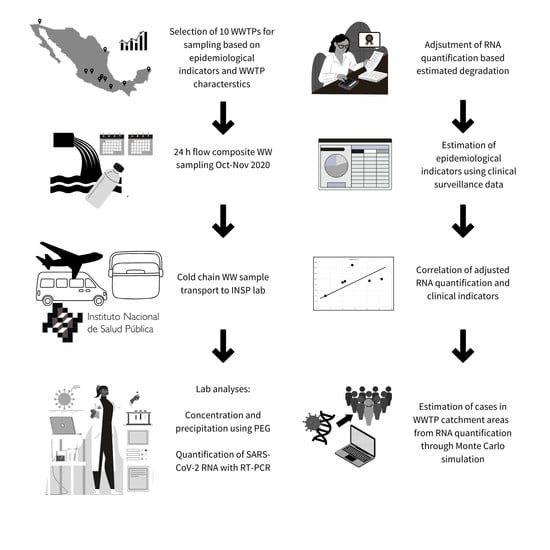
2. Materials and Methods
2.1. Instruments and Processes Standardization
2.2. Sampling Sites Selection
2.3. WW Sampling
2.4. Sample Concentration
2.5. Viral RNA Extraction, Detection, and Quantification
2.6. Recovery Efficiency Test
2.7. Clinical-Based Surveillance Data
2.8. Lead Time of WW-Based Surveillance
2.9. Infection Prevalence Estimation
3. Results
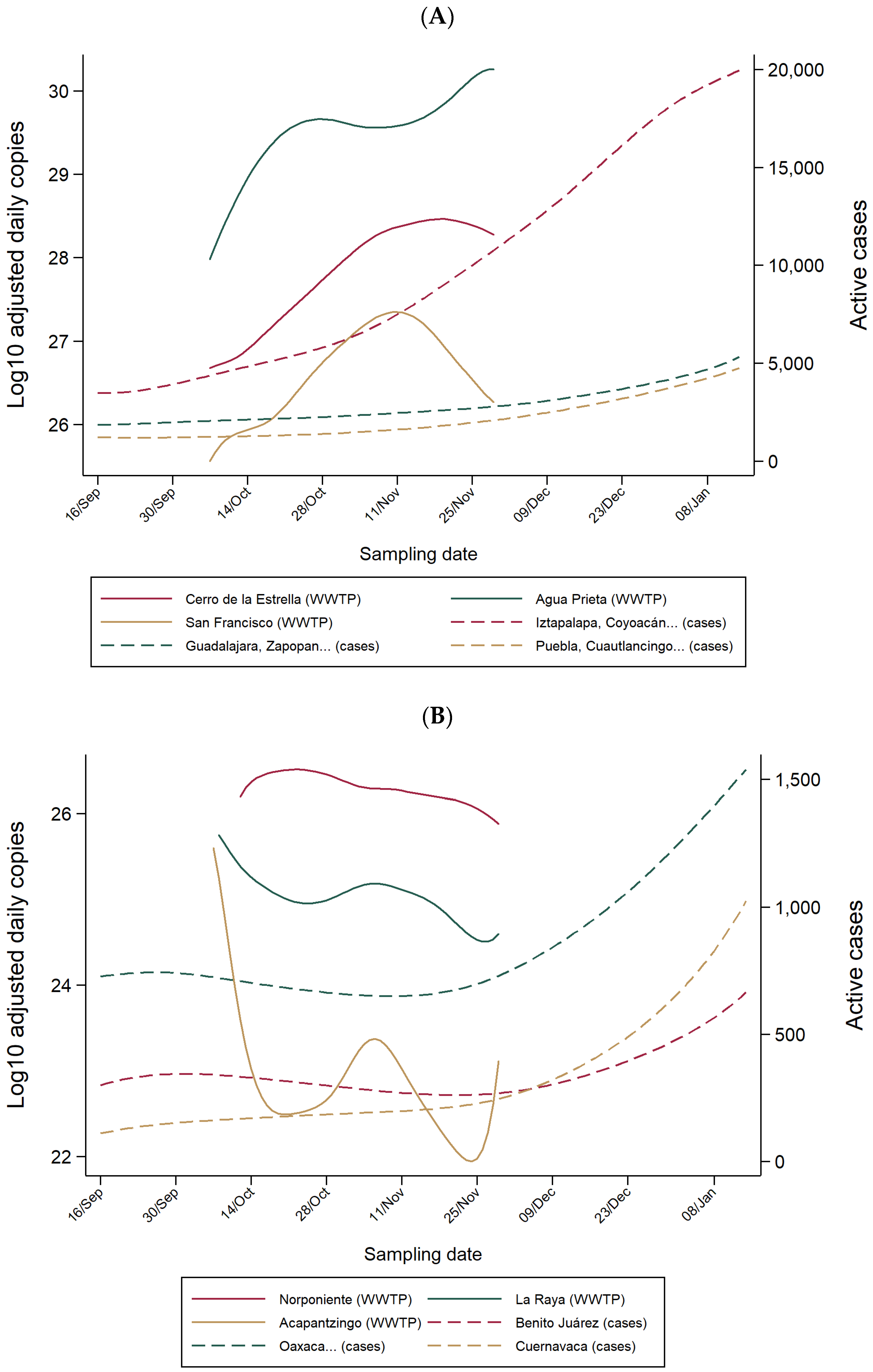
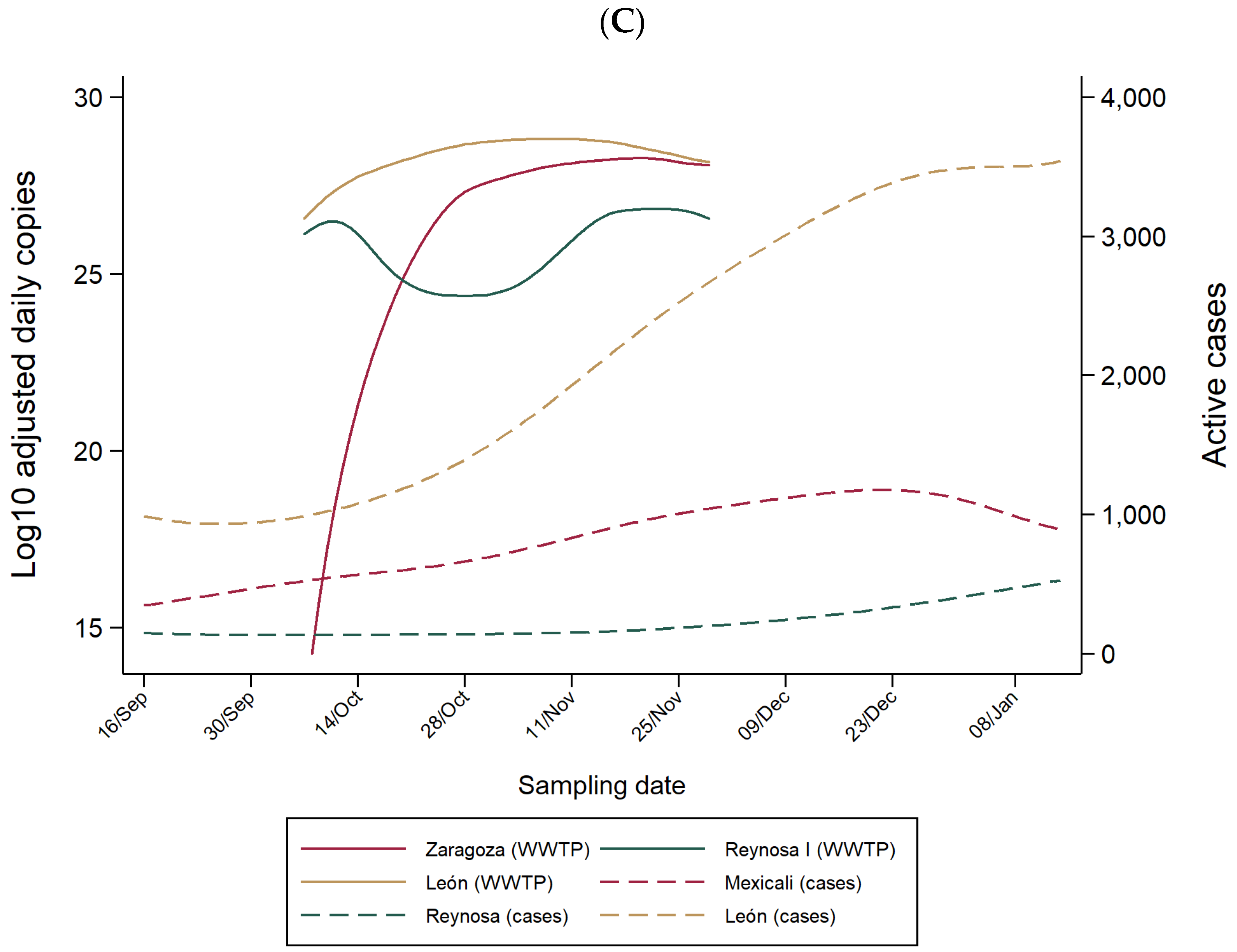
3.1. SARS-CoV-2 RNA in WW of WWTPs and COVID-19 Hospitals
3.2. Lead Time of WW-Based Surveillance Compared to Clinical-Based Surveillance
3.3. WW-Based COVID-19 Estimated Cases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Basto-Abreu, A.; Carnalla, M.; Torres-Ibarra, L.; Romero-Martínez, M.; Martínez-Barnetche, J.; López-Martínez, I.; Aparicio-Antonio, R.; Shamah-Levy, T.; Alpuche-Aranda, C.; Rivera, J.A.; et al. Nationally Representative SARS-CoV-2 Antibody Prevalence Estimates after the First Epidemic Wave in Mexico. Nat. Commun. 2022, 13, 589. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bello-Chavolla, O.Y.; Antonio-Villa, N.E.; Fernández-Chirino, L.; Guerra, E.C.; Fermín-Martínez, C.A.; Márquez-Salinas, A.; Vargas-Vázquez, A.; Bahena-López, J.P. Diagnostic Performance and Clinical Implications of Rapid SARS-CoV-2 Antigen Testing in Mexico Using Real-World Nationwide COVID-19 Registry Data. PLoS ONE 2021, 16, e0256447. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Luo, M.; Zou, Z.; Wang, X.; Chen, C.; Qiu, J. Asymptomatic SARS-CoV-2 Infected Case with Viral Detection Positive in Stool but Negative in Nasopharyngeal Samples Lasts for 42 Days. J. Med. Virol. 2020, 92, 1807–1809. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Y.; Shen, Y.; Shi, C.; Chen, Y.; Shi, P.; Gao, Y.; Wang, Y.; Lu, B. Detection of SARS-CoV-2 by RT-PCR in Anal from Patients Who Have Recovered from Coronavirus Disease 2019. J. Med. Virol. 2020, 92, 1769–1771. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged Presence of SARS-CoV-2 Viral RNA in Faecal Samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of Pediatric SARS-CoV-2 Infection and Potential Evidence for Persistent Fecal Viral Shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Zhang, Y.; Cen, M.; Hu, M.; Du, L.; Hu, W.; Kim, J.J.; Dai, N. Prevalence and Persistent Shedding of Fecal SARS-CoV-2 RNA in Patients with COVID-19 Infection: A Systematic Review and Meta-Analysis. Clin. Transl. Gastroenterol. 2021, 12, E00343. [Google Scholar] [CrossRef]
- Cruz-Cruz, C.; Rodríguez-Dozal, S.; Cortez-Lugo, M.; Ovilla-Muñoz, M.; Carnalla-Cortés, M.; Sánchez-Pájaro, A.; Schilmann, A. Revisión Rápida: Monitoreo de La Presencia e Infectividad Del Virus SARS-CoV-2 y Otros Coronavirus En Aguas Residuales. Salud Publica Mex. 2021, 63, 109–119. [Google Scholar] [CrossRef]
- Carrillo-Reyes, J.; Barragán-Trinidad, M.; Buitrón, G. Surveillance of SARS-CoV-2 in Sewage and Wastewater Treatment Plants in Mexico. J. Water Process Eng. 2020, 40, 101815. [Google Scholar] [CrossRef]
- González-Reyes, J.R.; Hernández-Flores, M.d.l.L.; Paredes-Zarco, J.E.; Téllez-Jurado, A.; Fayad-Meneses, O.; Carranza-Ramírez, L. Detection of SARS-CoV-2 in Wastewater Northeast of Mexico City: Strategy for Monitoring and Prevalence of COVID-19. Int. J. Environ. Res. Public Health 2021, 18, 8547. [Google Scholar] [CrossRef]
- Padilla-Reyes, D.A.; Álvarez, M.M.; Mora, A.; Cervantes-Avilés, P.A.; Kumar, M.; Loge, F.J.; Mahlknecht, J. Acquired Insights from the Long-Term Surveillance of SARS-CoV-2 RNA for COVID-19 Monitoring: The Case of Monterrey Metropolitan Area (Mexico). Environ. Res. 2022, 210, 112967. [Google Scholar] [CrossRef]
- Zarza, E.; Diego-García, E.; García, L.V.; Castro, R.; Mejía, G.; Herrera, D.; Cuevas, R.; Palomeque, Á.; Iša, P.; Guillén, K. Monitoring SARS-CoV-2 in the Wastewater and Rivers of Tapachula, a Migratory Hub in Southern Mexico. Food Environ. Virol. 2022, 14, 199–211. [Google Scholar] [CrossRef]
- Rosiles-González, G.; Carrillo-Jovel, V.H.; Alzate-Gaviria, L.; Betancourt, W.Q.; Gerba, C.P.; Moreno-Valenzuela, O.A.; Tapia-Tussell, R.; Hernández-Zepeda, C. Environmental Surveillance of SARS-CoV-2 RNA in Wastewater and Groundwater in Quintana Roo, Mexico. Food Environ. Virol. 2021, 13, 457–469. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y.; et al. Correlation of SARS-CoV-2 RNA in Wastewater with COVID-19 Disease Burden in Sewersheds. Sci. Total Environ. 2021, 775, 145790. [Google Scholar] [CrossRef]
- Giraud-Billoud, M.; Cuervo, P.; Altamirano, J.C.; Pizarro, M.; Aranibar, J.N.; Catapano, A.; Cuello, H.; Masachessi, G.; Vega, I.A. Monitoring of SARS-CoV-2 RNA in Wastewater as an Epidemiological Surveillance Tool in Mendoza, Argentina. Sci. Total Environ. 2021, 796, 148887. [Google Scholar] [CrossRef]
- Saguti, F.; Magnil, E.; Enache, L.; Churqui, M.P.; Johansson, A.; Lumley, D.; Davidsson, F.; Dotevall, L.; Mattsson, A.; Trybala, E.; et al. Surveillance of Wastewater Revealed Peaks of SARS-CoV-2 Preceding Those of Hospitalized Patients with COVID-19. Water Res. 2021, 189, 116620. [Google Scholar] [CrossRef]
- Kumar, M.; Joshi, M.; Patel, A.K.; Joshi, C.G. Unravelling the Early Warning Capability of Wastewater Surveillance for COVID-19: A Temporal Study on SARS-CoV-2 RNA Detection and Need for the Escalation. Environ. Res. 2021, 196, 110946. [Google Scholar] [CrossRef]
- D’Aoust, P.M.; Graber, T.E.; Mercier, E.; Montpetit, D.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Catching a Resurgence: Increase in SARS-CoV-2 Viral RNA Identified in Wastewater 48 h before COVID-19 Clinical Tests and 96 h before Hospitalizations. Sci. Total Environ. 2021, 770, 145319. [Google Scholar] [CrossRef]
- Farrell, J.A.; Whitmore, L.; Duffy, D.J. The Promise and Pitfalls of Environmental DNA and RNA Approaches for the Monitoring of Human and Animal Pathogens from Aquatic Sources. Bioscience 2021, 71, 609–625. [Google Scholar] [CrossRef]
- Kumar, M.; Jiang, G.; Kumar Thakur, A.; Chatterjee, S.; Bhattacharya, T.; Mohapatra, S.; Chaminda, T.; Kumar Tyagi, V.; Vithanage, M.; Bhattacharya, P.; et al. Lead Time of Early Warning by Wastewater Surveillance for COVID-19: Geographical Variations and Impacting Factors. Chem. Eng. J. 2022, 441, 135936. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.D.; Metcalf, T.G. Polyethylene Glycol Precipitation for Recovery of Pathogenic Viruses, Including Hepatitis A Virus and Human Rotavirus, from Oyster, Water, and Sediment Samples. Appl. Environ. Microbiol. 1988, 54, 1983–1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CDC. 2019-Novel Coronavirus (2019-NCoV) Real-Time RRT-PCR Panel Primers and Probes [WWW Document]; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020. [Google Scholar]
- Medema, G.; Been, F.; Heijnen, L.; Petterson, S. Implementation of Environmental Surveillance for SARS-CoV-2 Virus to Support Public Health Decisions: Opportunities and Challenges. Curr. Opin. Environ. Sci. Health 2020, 17, 49–71. [Google Scholar] [CrossRef] [PubMed]
- Robotto, A.; Lembo, D.; Quaglino, P.; Brizio, E.; Polato, D.; Civra, A.; Cusato, J.; Di Perri, G. Wastewater-Based SARS-CoV-2 Environmental Monitoring for Piedmont, Italy. Environ. Res. 2021, 203, 111901. [Google Scholar] [CrossRef]
- Ministry of Health Mexico. Lineamiento Estandarizado para la Vigilancia Epidemiológica y por Laboratorio de la Enfermedad Respiratoria Viral; Government of Mexico: Mexico City, Mexico, 2021.
- Li, X.; Kulandaivelu, J.; Zhang, S.; Shi, J.; Sivakumar, M.; Mueller, J.; Luby, S.; Ahmed, W.; Coin, L.; Jiang, G. Data-Driven Estimation of COVID-19 Community Prevalence through Wastewater-Based Epidemiology. Sci. Total Environ. 2021, 789, 147947. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational Analysis of SARS-CoV-2/COVID-19 Surveillance by Wastewater-Based Epidemiology Locally and Globally: Feasibility, Economy, Opportunities and Challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- Weiss, A.; Jellingsø, M.; Sommer, M.O.A. Spatial and Temporal Dynamics of SARS-CoV-2 in COVID-19 Patients: A Systematic Review and Meta-Analysis. EBioMedicine 2020, 58, 102916. [Google Scholar] [CrossRef]
- Rose, C.; Parker, A.; Jefferson, B.; Cartmell, E. The Characterization of Feces and Urine: A Review of the Literature to Inform Advanced Treatment Technology. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1827–1879. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.M.; Butler, D.; Orman, N.R.; Davies, J.W. Gross Solids Transport in Small Diameter Sewers. Water Sci. Technol. 1996, 33, 25–30. [Google Scholar] [CrossRef]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-Analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef]
- Van Nuijs, A.L.N.; Mougel, J.F.; Tarcomnicu, I.; Bervoets, L.; Blust, R.; Jorens, P.G.; Neels, H.; Covaci, A. Sewage Epidemiology—A Real-Time Approach to Estimate the Consumption of Illicit Drugs in Brussels, Belgium. Environ. Int. 2011, 37, 612–621. [Google Scholar] [CrossRef]
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M.; et al. Measurement of SARS-CoV-2 RNA in Wastewater Tracks Community Infection Dynamics. Nat. Biotechnol. 2020, 38, 1164–1167. [Google Scholar] [CrossRef]
- Olesen, S.W.; Imakaev, M.; Duvallet, C. Making Waves: Defining the Lead Time of Wastewater-Based Epidemiology for COVID-19. Water Res. 2021, 202, 117433. [Google Scholar] [CrossRef]
- Bibby, K.; Bivins, A.; Wu, Z.; North, D. Making Waves: Plausible Lead Time for Wastewater Based Epidemiology as an Early Warning System for COVID-19. Water Res. 2021, 202, 117438. [Google Scholar] [CrossRef]
- Maal-Bared, R.; Qiu, Y.; Li, Q.; Gao, T.; Hrudey, S.E.; Bhavanam, S.; Ruecker, N.J.; Ellehoj, E.; Lee, B.E.; Pang, X. Does Normalization of SARS-CoV-2 Concentrations by Pepper Mild Mottle Virus Improve Correlations and Lead Time between Wastewater Surveillance and Clinical Data in Alberta (Canada): Comparing Twelve SARS-CoV-2 Normalization Approaches. Sci. Total Environ. 2023, 856, 158964. [Google Scholar] [CrossRef]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Thompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 Surveillance in Southeastern Virginia Using Wastewater-Based Epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef]
- Ahmed, W.; Tscharke, B.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Choi, P.; Clarke, L.; Dwyer, J.; Edson, J.; Nguyen, T.M.H.; et al. SARS-CoV-2 RNA Monitoring in Wastewater as a Potential Early Warning System for COVID-19 Transmission in the Community: A Temporal Case Study. Sci. Total Environ. 2021, 761, 144216. [Google Scholar] [CrossRef]
- Fongaro, G.; Stoco, P.H.; Souza, D.S.M.; Grisard, E.C.; Magri, M.E.; Rogovski, P.; Schörner, M.A.; Barazzetti, F.H.; Christoff, A.P.; de Oliveira, L.F.V.; et al. The Presence of SARS-CoV-2 RNA in Human Sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021, 778, 146198. [Google Scholar] [CrossRef]
- Rusiñol, M.; Zammit, I.; Itarte, M.; Forés, E.; Martínez-Puchol, S.; Girones, R.; Borrego, C.; Corominas, L.; Bofill-Mas, S. Monitoring Waves of the COVID-19 Pandemic: Inferences from WWTPs of Different Sizes. Sci. Total Environ. 2021, 787, 147463. [Google Scholar] [CrossRef] [PubMed]
- Sakarovitch, C.; Schlosser, O.; Courtois, S.; Proust-Lima, C.; Couallier, J.; Pétrau, A.; Litrico, X.; Loret, J.-F. Monitoring of SARS-CoV-2 in Wastewater: What Normalisation for Improved Understanding of Epidemic Trends? J. Water Health 2022, 20, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Oishi, W.; Maruo, C.; Saito, M.; Chen, R.; Kitajima, M.; Sano, D. Early Warning of COVID-19 via Wastewater-Based Epidemiology: Potential and Bottlenecks. Sci. Total Environ. 2021, 767, 145124. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.J.; Lo Jacomo, A.; Armenise, E.; Brown, M.R.; Bunce, J.T.; Cameron, G.J.; Fang, Z.; Farkas, K.; Gilpin, D.F.; Graham, D.W.; et al. Understanding and Managing Uncertainty and Variability for Wastewater Monitoring beyond the Pandemic: Lessons Learned from the United Kingdom National COVID-19 Surveillance Programmes. J. Hazard. Mater. 2022, 424, 127456. [Google Scholar] [CrossRef]
- Islam, G.; Gedge, A.; Lara-Jacobo, L.; Kirkwood, A.; Simmons, D.; Desaulniers, J.P. Pasteurization, Storage Conditions and Viral Concentration Methods Influence RT-QPCR Detection of SARS-CoV-2 RNA in Wastewater. Sci. Total Environ. 2022, 821, 153228. [Google Scholar] [CrossRef]
- Markt, R.; Mayr, M.; Peer, E.; Wagner, A.O.; Lackner, N.; Insam, H. Detection and Stability of SARS-CoV-2 Fragments in Wastewater: Impact of Storage Temperature. Pathogens 2021, 10, 1215. [Google Scholar] [CrossRef]
- Palmer, E.J.; Maestre, J.P.; Jarma, D.; Lu, A.; Willmann, E.; Kinney, K.A.; Kirisits, M.J. Development of a Reproducible Method for Monitoring SARS-CoV-2 in Wastewater. Sci. Total Environ. 2021, 799, 149405. [Google Scholar] [CrossRef]
- Pecson, B.M.; Darby, E.; Haas, C.N.; Amha, Y.M.; Bartolo, M.; Danielson, R.; Dearborn, Y.; Di Giovanni, G.; Ferguson, C.; Fevig, S.; et al. Reproducibility and Sensitivity of 36 Methods to Quantify the SARS-CoV-2 Genetic Signal in Raw Wastewater: Findings from an Interlaboratory Methods Evaluation in the U.S. Environ. Sci. Water Res. Technol. 2021, 7, 504–520. [Google Scholar] [CrossRef]
- Robinson, C.A.; Hsieh, H.Y.; Hsu, S.Y.; Wang, Y.; Salcedo, B.T.; Belenchia, A.; Klutts, J.; Zemmer, S.; Reynolds, M.; Semkiw, E.; et al. Defining Biological and Biophysical Properties of SARS-CoV-2 Genetic Material in Wastewater. Sci. Total Environ. 2022, 807, 150786. [Google Scholar] [CrossRef]
- Ampuero, M.; Valenzuela, S.; Valiente-Echeverría, F.; Soto-Rifo, R.; Barriga, G.; Chnaiderman, J.; Rojas, C.; Guajardo-Leiva, S.; Díez, B.; Gaggero, A. SARS-CoV-2 Detection in Sewage in Santiago, Chile—Preliminary Results. medRxiv 2020. [Google Scholar] [CrossRef]
- Achak, M.; Alaoui Bakri, S.; Chhiti, Y.; M’hamdi Alaoui, F.E.; Barka, N.; Boumya, W. SARS-CoV-2 in Hospital Wastewater during Outbreak of COVID-19: A Review on Detection, Survival and Disinfection Technologies. Sci. Total Environ. 2021, 761, 143192. [Google Scholar] [CrossRef]
- Wang, X.W.; Li, J.; Guo, T.; Zhen, B.; Kong, Q.; Yi, B.; Li, Z.; Song, N.; Jin, M.; Xiao, W.; et al. Concentration and Detection of SARS Coronavirus in Sewage from Xiao Tang Shan Hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Sci. Technol. 2005, 52, 213–221. [Google Scholar] [CrossRef]
- Chavarria-Miró, G.; Anfruns-Estrada, E.; Martínez-Velázquez, A.; Vázquez-Portero, M.; Guix, S.; Paraira, M.; Galofré, B.; Sánchez, G.; Pintó, R.M.; Bosch, A. Time Evolution of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Wastewater during the First Pandemic Wave of COVID-19 in the Metropolitan Area of Barcelona, Spain. Appl. Environ. Microbiol. 2021, 87, e02750-20. [Google Scholar] [CrossRef]
- Gonçalves, J.; Koritnik, T.; Mioč, V.; Trkov, M.; Bolješič, M.; Berginc, N.; Prosenc, K.; Kotar, T.; Paragi, M. Detection of SARS-CoV-2 RNA in Hospital Wastewater from a Low COVID-19 Disease Prevalence Area. Sci. Total Environ. 2021, 755, 4–10. [Google Scholar] [CrossRef]
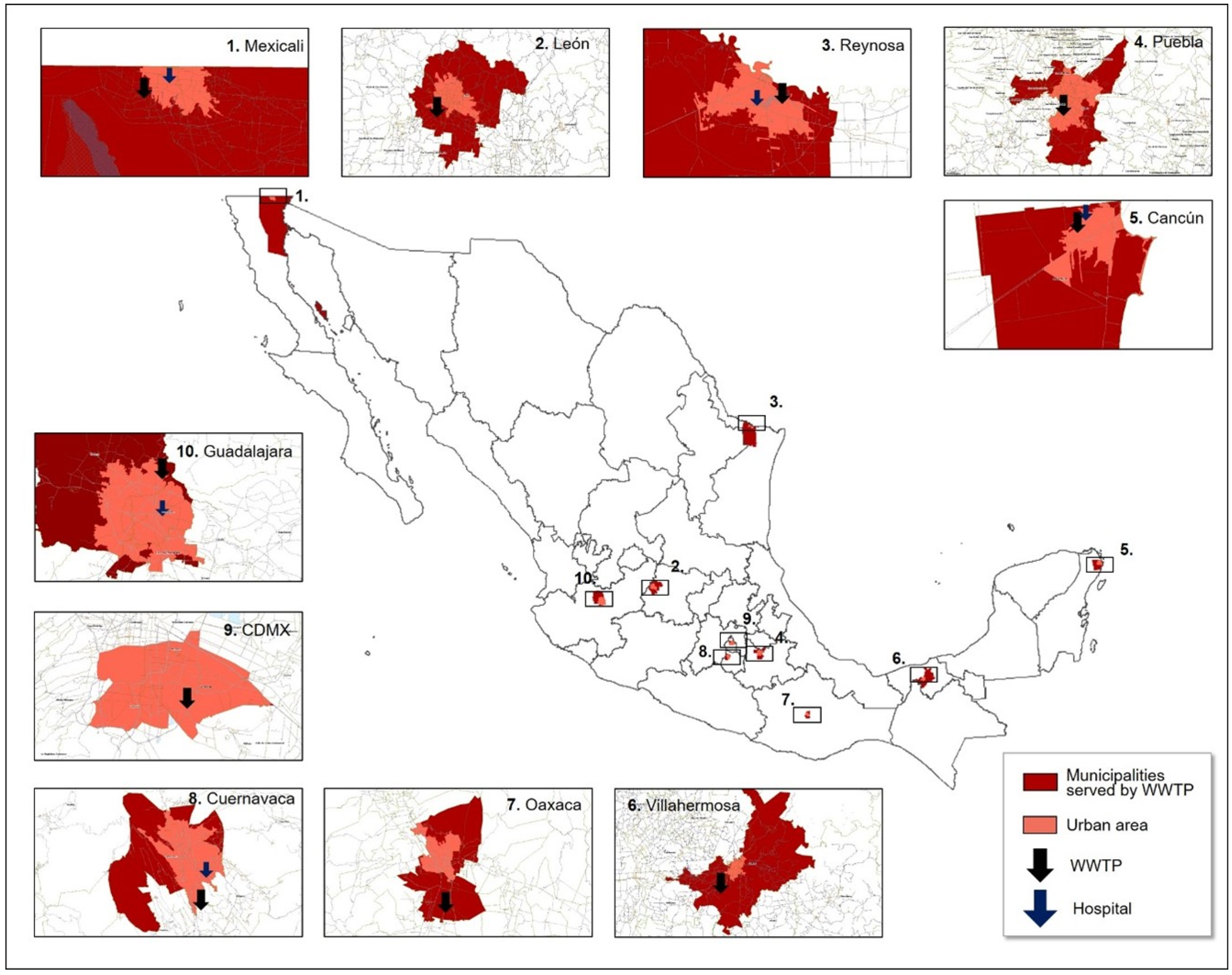

| City, State | WWTP | Municipalities Served by WWTP | RNA Copies/mL Median (min; max) | Average Daily Flow (L/s), Range | Average Daily Temperature (°C), Range | Average Distance Traveled (km) | Sewage System and WW Characteristics | Sampled COVID-19 Hospital | Hospital RNA Copies/mL Median (min; max) | Reported Hospital WW Chlorination |
|---|---|---|---|---|---|---|---|---|---|---|
| Guadalajara, Jalisco | Agua Prieta (AP) | Zapopan, Guadalajara, Tlaquepaque | 542 (114.6; 4396.8) | 4599 (4009, 5330) | 23.7 (22.0, 25.3) | 8.3 | No data | Hospital 1 | 209.3 (0; 32,460) | No |
| Mexico City, Mexico City | Cerro de la Estrella (CE) | Iztapalapa, Iztacalco, Benito Juárez, Coyoacán | 377.2 (84.3; 933.8) | 1867 (1303, 2505) | 19.3 (17.7, 20.3) | 16.5 | Only domestic discharges | NS | NS | NS |
| Puebla, Puebla | San Francisco (SF) | Puebla, Cuautlancingo, San Pedro Cholula | 150.9 (54.1; 2379.3) | 1107 (985, 1219) | 21.0 (19.3, 22.0) | 5.6 | Industrial, chemical, and tourism discharges 6% Runoffs | NS | NS | NS |
| Mexicali, Baja California | Zaragoza (ZA) | Mexicali | 616.8 (0; 2037.8) | 882 (693, 1303) | 27.7 (23.9, 30.8) | 3.3 | Industrial, chemical, and tourism discharges | Hospital 2 | 1240.5 (0; 21,223.2) | No |
| León, Guanajuato | León (LE) | León | 659.3 (66.3; 1378.5) | 522 (166, 1017) | 24.5 (22.8, 26.1) | 6.4 | Industrial, chemical, and tourism discharges Runoffs | NS | NS | NS |
| Reynosa, Tamaulipas | Reynosa I (RE) | Reynosa | 128.6 (0; 451.8) | 488 (480, 495) | 26.6 (25.1, 28.4) | 6.9 | Industrial, chemical, and tourism discharges 17% Runoffs | Hospital 3 | 0 (0; 74.2) | Yes |
| Cancún, Quintana Roo | Norponiente (NO) | Benito Juárez | 159.9 (56.7; 423.7) | 216 (123, 273) | 29.1 (27.7, 30.3) | 2.5 | Only domestic discharges | Hospital 4 | 0 (0; 0) | Yes |
| Cuernavaca, Morelos | Acapantzingo (AC) | Cuernavaca | 39.7 (0; 181.2) | 259 (225, 316) | 20.4 (19.1, 21.4) | 4.1 | Industrial and tourism discharges | Hospital 5 | 3393.6 (195.9; 94,936.3) | No |
| Oaxaca, Oaxaca | La Raya (LR) | Santa Cruz Xoxocotlán, Ánimas Trujano, San Agustín de las Juntas, San Antonio de la Cal, Santa María Coyotepec, Oaxaca de Juárez, Santa Lucía del Camino, San Raymundo Jalpan, San Simón Almolongas, San Agustín Yatareni, San Andrés Huayápam, San Jacinto Amilpas | 322.9 (74.7; 646.6) | 33 (28, 38) | 23.6 (22.7, 24.2) | 9.5 | Industrial and tourism discharges 80% Runoffs | NS | NS | NS |
| Villahermosa, Tabasco | Zona Noreste | Centro | 0 (0,0) | 235 (12, 250) | 26.7 (24.6, 28.1) | 2 | Only domestic discharges 15% Runoffs | NS | NS | NS |
| Active Cases | |||
|---|---|---|---|
| WWTP | City, State | Lag (Days) | Maximum Rho |
| All sites | 39 | 0.66 | |
| AP | Guadalajara, Jalisco | 2 37 | 0.71 0.62 |
| CE | Mexico City, Mexico City | 1 39 | 0.82 0.91 |
| SF | Puebla, Puebla | 35 | 0.44 |
| ZA | Mexicali, Baja California | 0 40 | 0.39 0.59 |
| LE | León, Guanajuato | 40 | 0.75 |
| RE | Reynosa, Tamaulipas | 18 43 | 0.52 0.33 |
| Wastewater (WW) Surveillance | Clinical Surveillance at Municipal Level | WW/Municipal Ratios | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WWTP | City, State | Testing Rate per 100,000 | Estimated Cases Median (min; max) | Estimated Catchment Area Population | Estimated Prevalence per 1000 People Median (min; max) | Active Cases Median (min; max) | Municipal Population | Active Cases Prevalence per 1000 People Median (min; max) | Estimated Cases/Active Cases Ratio Median (min; max) | Estimated Prevalence/Active Cases Prevalence Ratio Median (min; max) |
| AP | Guadalajara, Jalisco | 1528 (Zapopan) | 53,295 (10,533; 423,940) | 1,454,428 | 36.6 (7.2; 291.5) | 1993 (1673; 2493) | 3,456,613 | 0.6 (0.5; 0.7) | 24.8 (6.3; 192.4) | 58.9 (15.0; 457.1) |
| CE | Mexico City, Mexico City | 5796 (Iztapalapa) | 12,293 (2342; 26,321) | 172,610 | 71.2 (13.6; 152.5) | 5317 (3274; 5782) | 3,244,111 | 1.6 (1.0; 1.8) | 2.4 (0.6; 5.4) | 45.7 (10.9; 102.0) |
| SF | Puebla, Puebla | 3829 (Puebla) | 2779 (1024; 45,157) | 321,597 | 8.6 (3.2; 140.4) | 1181 (993; 1754) | 2,561,142 | 0.5 (0.4; 0.7) | 2.4 (0.9; 39.7) | 18.8 (6.9; 316.0) |
| ZA | Mexicali, Baja California | 3026 | 9698 (1940; 29,811) | 267,815 | 36.2 (7.2; 111.3) | 543 (450; 1067) | 988,417 | 0.5 (0.5; 1.1) | 16.4 (3.4; 66.2) | 60.6 (12.6; 244.5) |
| LE | León, Guanajuato | 3462 | 10,240 (1264; 21,845) | 428,684 | 23.9 (2.9; 51.0) | 1284 (788; 2669) | 1,578,626 | 0.8 (0.5; 1.7) | 7.8 (1.6; 19.6) | 28.8 (5.9; 72.2) |
| RE | Reynosa, Tamaulipas | 2193 | 2129 (471; 6870) | 117,471 | 18.1 (4.0; 58.5) | 114 (81; 153) | 646,202 | 0.2 (0.1; 0.2) | 20.9 (3.8; 56.7) | 115.0 (20.9; 312.0) |
| NO | Cancún, Quintana Roo | 1730 | 1066 (420; 3032) | 148,180 | 7.2 (2.8; 20.5) | 234 (153; 287) | 743,626 | 0.3 (0.2; 0.4) | 5.5 (1.7; 13.8) | 27.8 (8.6; 69.4) |
| AC | Cuernavaca, Morelos | 2322 | 262 (83; 802) | 16,335 | 16.1 (5.1; 49.1) | 139 (103; 166) | 366,321 | 0.4 (0.3; 0.5) | 2.0 (0.6; 5.4) | 45.9 (13.5; 121.5) |
| LR | Oaxaca, Oaxaca | 7238 | 1846 (405; 3840) | 144,707 | 12.8 (2.8; 26.5) | 548 (465; 717) | 477,712 | 1.1 (1.0; 1.5) | 3.2 (0.8; 7.4) | 10.4 (2.5; 24.4) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schilmann, A.; Sánchez-Pájaro, A.; Ovilla-Muñoz, M.T.; Téllez-Sosa, J.; Bravo-Romero, S.; Bahena-Reyes, S.Y.; Lobato, M.; Martínez-Barnetche, J.; Alpuche-Aranda, C.M.; Lamadrid-Figueroa, H.; et al. SARS-CoV-2 Wastewater Surveillance in Ten Cities from Mexico. Water 2023, 15, 799. https://doi.org/10.3390/w15040799
Schilmann A, Sánchez-Pájaro A, Ovilla-Muñoz MT, Téllez-Sosa J, Bravo-Romero S, Bahena-Reyes SY, Lobato M, Martínez-Barnetche J, Alpuche-Aranda CM, Lamadrid-Figueroa H, et al. SARS-CoV-2 Wastewater Surveillance in Ten Cities from Mexico. Water. 2023; 15(4):799. https://doi.org/10.3390/w15040799
Chicago/Turabian StyleSchilmann, Astrid, Andrés Sánchez-Pájaro, Marbella T. Ovilla-Muñoz, Juan Téllez-Sosa, Sugey Bravo-Romero, Sara Yuvisela Bahena-Reyes, Margarita Lobato, Jesús Martínez-Barnetche, Celia Mercedes Alpuche-Aranda, Héctor Lamadrid-Figueroa, and et al. 2023. "SARS-CoV-2 Wastewater Surveillance in Ten Cities from Mexico" Water 15, no. 4: 799. https://doi.org/10.3390/w15040799