Risks of Antibiotic Resistance Dissemination by Leachates from Municipal Landfills of Different Ages
Abstract
:1. Introduction
2. Materials and Methods
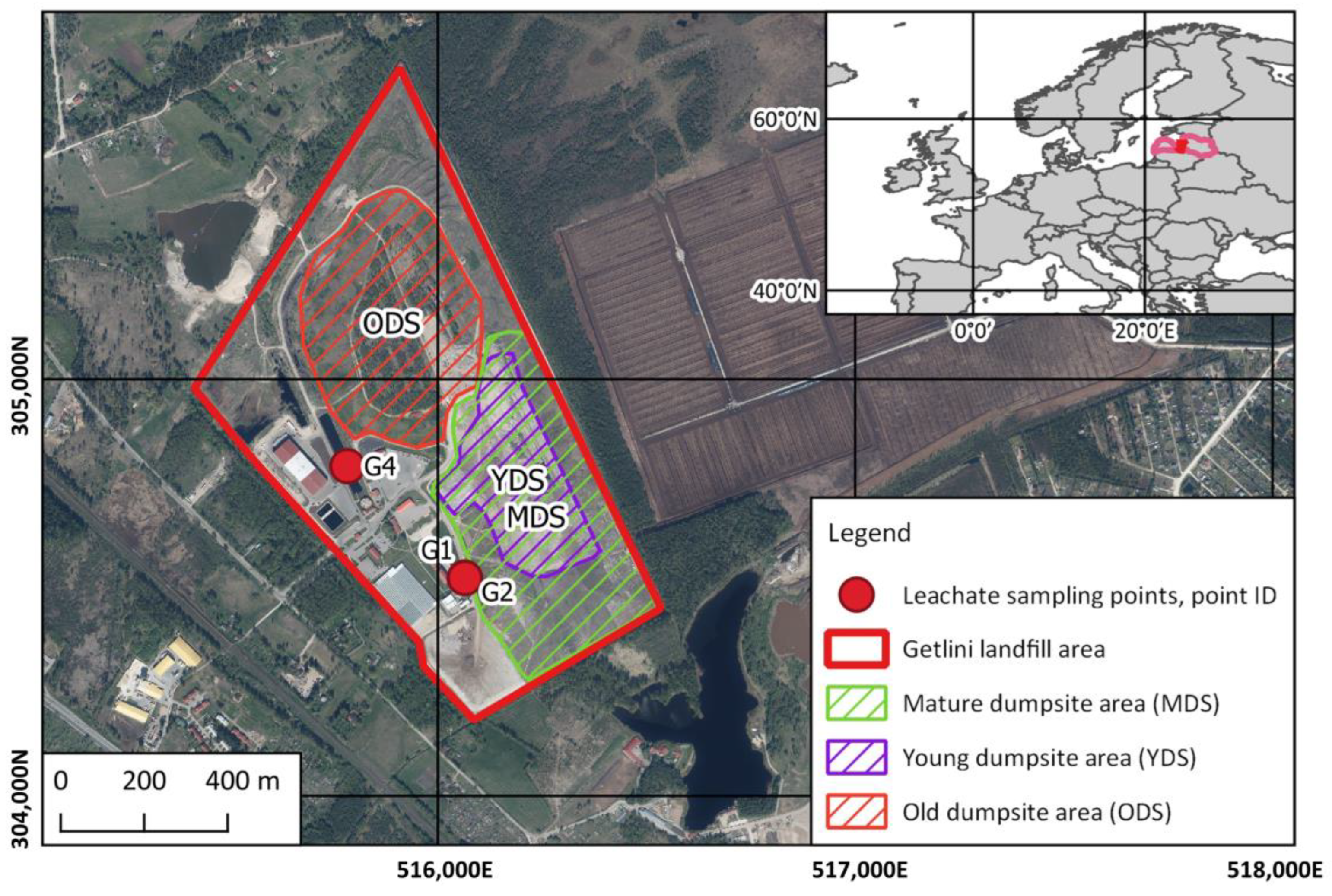
2.1. Characterization of the Getlini Landfill and Leachate Collection
2.2. Physicochemical Testing of Leachates
2.3. Analytical Determination of Micropollutants
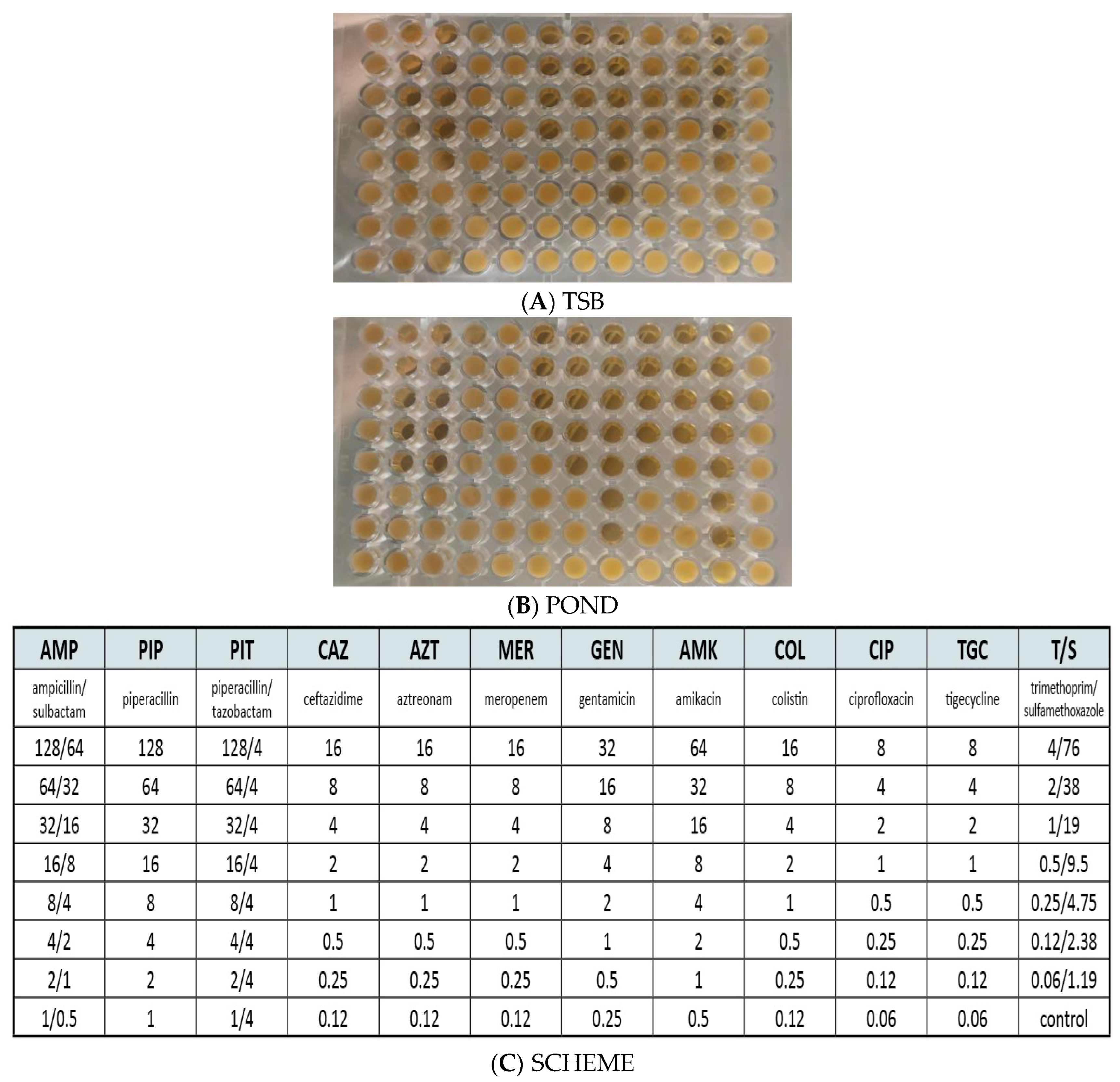
2.4. Characterization of Culturable Microorganisms
2.5. Testing of Microbial DNA in Leachates
2.5.1. DNA Extraction and Shotgun Sequencing
2.5.2. Shotgun Sequencing Data Analysis
2.5.3. Identification of Microbial Resistance Genes
2.6. Risk of AMR Induction by Leachates
2.7. Microscopy Study
2.8. Statistical Analysis
3. Results
3.1. Physicochemical Characterization of Leachates
3.2. Analytical Testing of Micropollutants
3.3. Characterization of Culturable Microorganisms
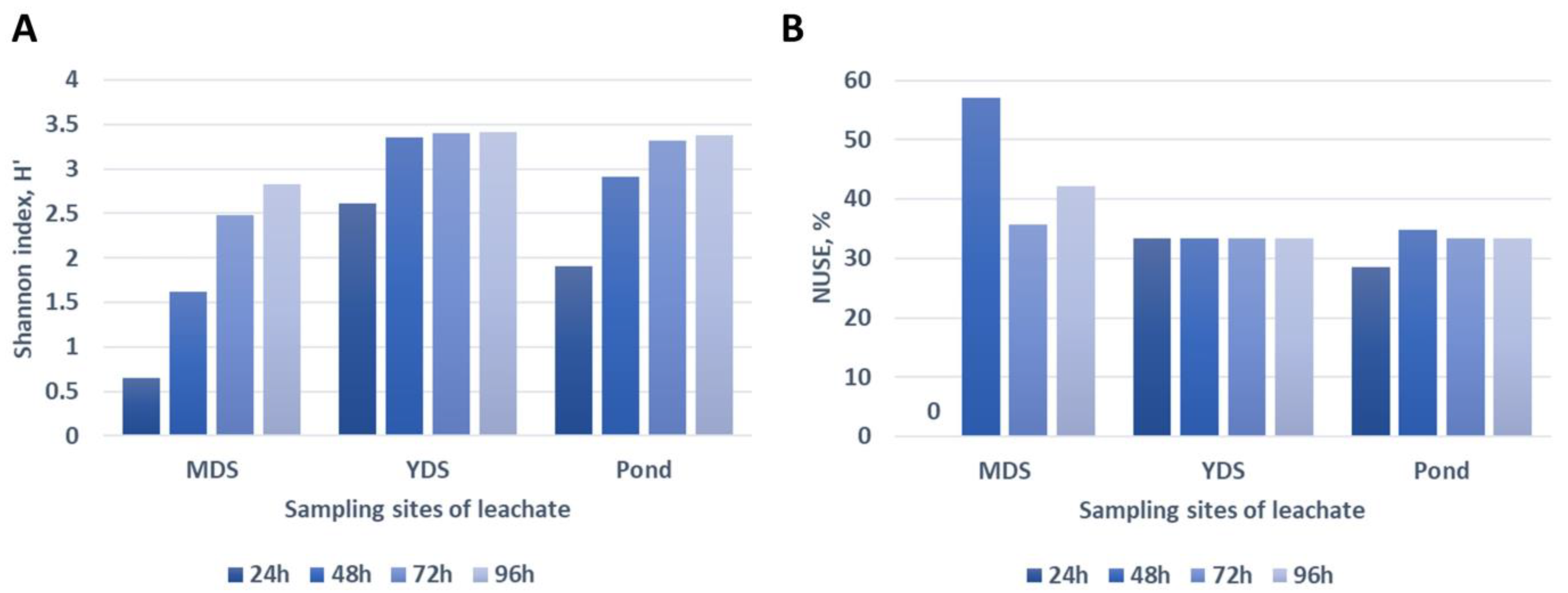
3.4. Taxonomic Profile of Leachate Samples
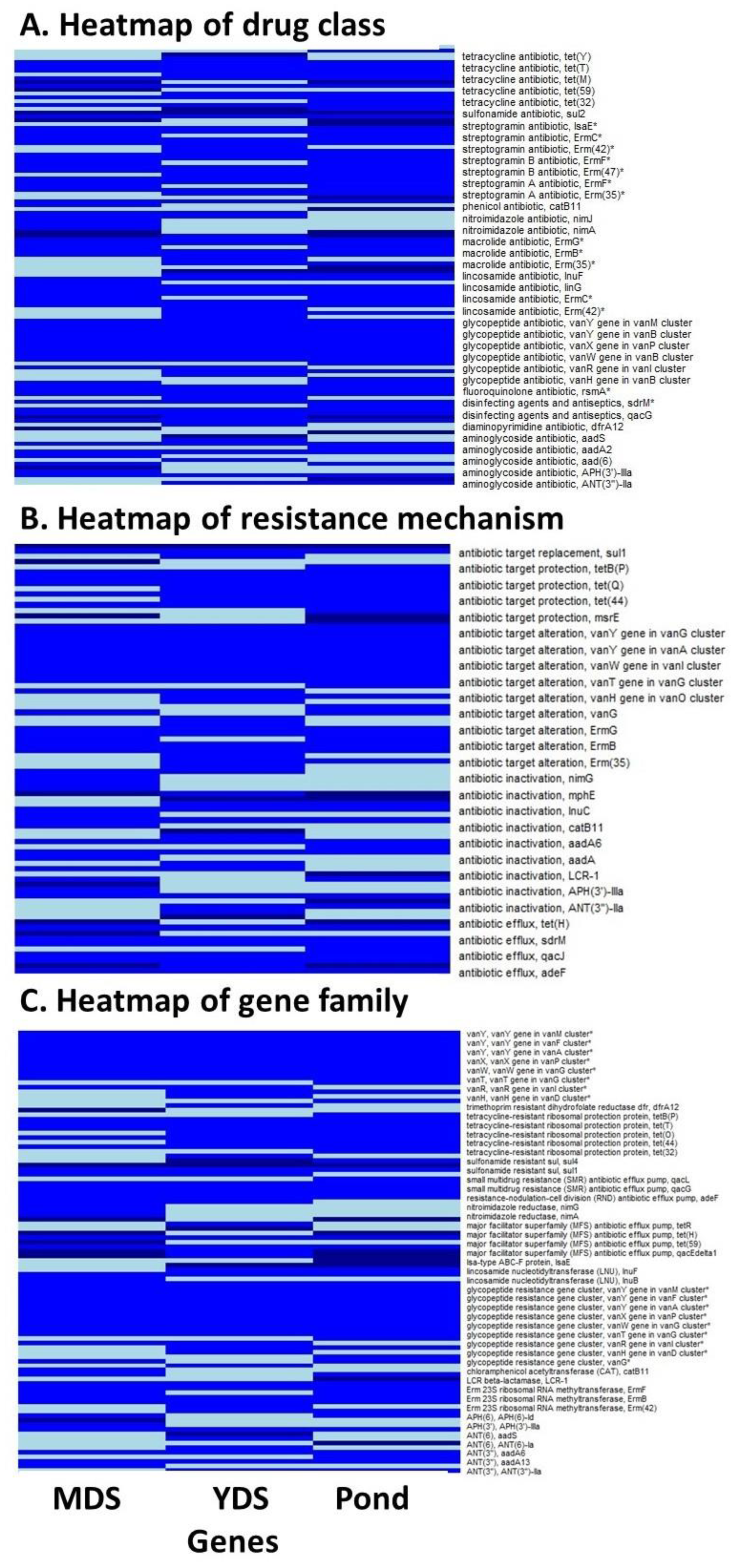
Antimicrobial Resistance Genes in Metagenomic Data
3.5. Risk of AMR Induction by Leachates
4. Discussion
5. Conclusions
- Metagenomic analysis showed a prevalence of Proteobacteria (43.76–54.70%), followed by Firmicutes (8.51–13.11%) and Euryarchaeota (9.37–12.53%) in all three leachate samples. Regarding the inner diversity of samples, the highest Chao1 measure was obtained for YDS—8885.95, followed by MDS and pond, whereas the richness estimator, e.g., Shannon index, was the highest for pond—7.23, followed by YDS and MDS. Similarly, the highest value of the evenness estimator—the Simpson index—was observed for the pond—0.99, followed by YDS and MDS.
- The number and composition of specific ARGs found in leachate samples derived from MDS, YDS, and ponds slightly varied. In total, 80 ARGs belonging to 31 distinct ARG families were identified. These ARGs are known to confer resistance against 19 different classes of antibiotics, such as aminoglycosides, glycopeptides, fluoroquinolones, lincosamides, macrolides, streptogramins, and tetracyclines, as well as disinfecting agents and antiseptics.
- The incubation of P. putida MSCL650 in sterilized leachate derived from the pond resulted in a decrease in MIC for several antibiotics compared to the control. This effect can be explained by the high concentration of toxic compounds in leachates. Leachates from MDS and YDS totally inhibited the cells of P. putida.
- A comparison of two dumping sites tested in the study, which differed by waste composition and treatment and the duration of operations, did not reveal considerable differences in the microbial community structure and ARG abundance. The lower ecotoxicity in the leachate from the pond compared to those from MDS and YDS pointed to the effectiveness of solid waste treatment.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Priority Group | Relative Abundance *, % | ||
|---|---|---|---|
| 1. Critical | MDS | YDS | Pond |
| Acinetobacter baumannii, carbapenem-resistant | 0.046 | 0.034 | 0.037 |
| Pseudomonas aeruginosa, carbapenem-resistant | 0.233 | 0.174 | 0.222 |
| Enterobacteriaceae, carbapenem-resistant, ESBL-producing | 1.294 | 0.984 | 1.324 |
| 2. High | |||
| Enterococcus faecium, vancomycin-resistant | 0.015 | 0.012 | 0.020 |
| Salmonellae, fluoroquinolone-resistant | n.d. | n.d. | n.d. |
| Staphylococcus aureus, methicillin-resistant, vancomycin-intermediate and resistant | n.d. | n.d. | n.d. |
| Helicobacter pylori, clarithromycin-resistant | 0.032 | 0.049 | 0.032 |
| Campylobacter spp., fluoroquinolone-resistant | 0.470 | 1.305 | 0.385 |
| Neisseria gonorrhoeae, cephalosporin-resistant, fluoroquinolone-resistant | 0.001 | 0.001 | 0.002 |
| 3. Medium | |||
| Streptococcus pneumoniae, penicillin-non-susceptible | 0.011 | 0.009 | 0.009 |
| Haemophilus influenzae, ampicillin-resistant | n.d. | n.d. | n.d. |
| Shigella spp., fluoroquinolone-resistant | 0.002 | 0.004 | 0.005 |
| Name | MDS | YDS | Pond | Lineage |
|---|---|---|---|---|
| Pandoravirus neocaledonia | 40 | 89 | 267 | Viruses > unclassified viruses > unclassified DNA viruses > unclassified dsDNA viruses > Pandoravirus |
| Alteromonas phage vB_AmaP_AD45-P1 | 13 | 159 | 193 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Podoviridae > unclassified Podoviridae |
| Aeromonas phage phiAS5 | 21 | 5 | 173 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Myoviridae > Tevenvirinae > unclassified Tevenvirinae |
| Brevibacillus phage Sundance | 15 | 66 | 162 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Siphoviridae > unclassified Siphoviridae |
| Yersinia virus phiR201 | 11 | 39 | 149 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Demerecviridae > Markadamsvirinae > Haartmanvirus |
| Halovirus HRTV-8 | 3 | 37 | 145 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Myoviridae > unclassified Myoviridae |
| Paramecium bursaria Chlorella virus AR158 | 18 | 45 | 131 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Algavirales > Phycodnaviridae > Chlorovirus > unclassified Chlorovirus |
| Megavirus chiliensis | 94 | 100 | 91 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Imitervirales > Mimiviridae > Mimivirus > unclassified Mimivirus |
| Orpheovirus IHUMI-LCC2 | 81 | 150 | 80 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Algavirales > Phycodnaviridae > unclassified Phycodnaviridae |
| Moumouvirus | 116 | 240 | 63 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Imitervirales > Mimiviridae > unclassified Mimiviridae |
| Cafeteria roenbergensis virus | 102 | 127 | 63 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Imitervirales > Mimiviridae > Cafeteriavirus |
| Lymphocystis disease virus—isolate China | 106 | 31 | 46 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Pimascovirales > Iridoviridae > Alphairidovirinae > Lymphocystivirus > unclassified Lymphocystivirus |
| Bacillus virus G | 95 | 129 | 45 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Myoviridae |
| Aureococcus anophagefferens virus | 48 | 145 | 38 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Algavirales > Phycodnaviridae > unclassified Phycodnaviridae |
| Chrysochromulina ericina virus | 25 | 142 | 37 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Algavirales > Phycodnaviridae > unclassified Phycodnaviridae |
| Mimivirus terra2 | 72 | 109 | 26 | Viruses > Varidnaviria > Bamfordvirae > Nucleocytoviricota > Megaviricetes > Imitervirales > Mimiviridae > Mimivirus > unclassified Mimivirus |
| Escherichia virus JSE | 111 | 0 | 1 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Myoviridae > Tevenvirinae > Krischvirus |
| Arthrobacter virus Coral | 121 | 0 | 0 | Viruses > Duplodnaviria > Heunggongvirae > Uroviricota > Caudoviricetes > Caudovirales > Siphoviridae > Coralvirus |

References
- Nava, A.R.; Daneshian, L.; Sarma, H. Antibiotic Resistant Genes in the Environment-Exploring Surveillance Methods and Sustainable Remediation Strategies of Antibiotics and ARGs. Environ. Res. 2022, 215, 114–212. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Xiong, J.-Q.; Patil, S.M.; Ha, G.-S.; Hoh, J.-K.; Park, H.-K.; Chung, W.; Chang, S.W.; Khan, M.A.; Park, H.B.; et al. Dissemination of Sulfonamide Resistance Genes in Digester Microbiome during Anaerobic Digestion of Food Waste Leachate. J. Hazard. Mater. 2023, 452, 131200. [Google Scholar] [CrossRef]
- Sun, H.; Bjerketorp, J.; Levenfors, J.J.; Schnürer, A. Isolation of Antibiotic-Resistant Bacteria in Biogas Digestate and Their Susceptibility to Antibiotics. Environ. Pollut. 2020, 266, 115265. [Google Scholar] [CrossRef]
- European Commission. A European One Health Action Plan against Antimicrobial Resistance (AMR); European Commission: Brussels, Belgium, 2017. [Google Scholar]
- Taing, L.; Bhatia, H.; Kaiser, R.A.; Qadir, M.; Mehmood, H. A Rapid Review of Environmental Health Gaps in Antimicrobial Resistance and Water-Related Research from 1990–2020. Int. J. Environ. Res. Public Health 2022, 19, 6549. [Google Scholar] [CrossRef] [PubMed]
- Liguori, K.; Keenum, I.; Davis, B.C.; Calarco, J.; Milligan, E.; Harwood, V.J.; Pruden, A. Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control. Environ. Sci. Technol. 2022, 56, 9149–9160. [Google Scholar] [CrossRef] [PubMed]
- Burch, T.R.; Newton, R.J.; Kimbell, L.K.; LaMartina, E.L.; O’Malley, K.; Thomson, S.M.; Marshall, C.W.; McNamara, P.J. Targeting Current and Future Threats: Recent Methodological Trends in Environmental Antimicrobial Resistance Research and Their Relationships to Risk Assessment. Environ. Sci. 2022, 8, 1787–1802. [Google Scholar] [CrossRef]
- Irfan, M.; Almotiri, A.; AlZeyadi, Z.A. Antimicrobial Resistance and Its Drivers—A Review. Antibiotics 2022, 11, 362. [Google Scholar] [CrossRef]
- Bai, Y.; Xu, R.; Wang, Q.P.; Zhang, Y.R.; Yang, Z.H. Sludge Anaerobic Digestion with High Concentrations of Tetracyclines and Sulfonamides: Dynamics of Microbial Communities and Change of Antibiotic Resistance Genes. Bioresour. Technol. 2019, 276, 51–59. [Google Scholar] [CrossRef]
- Ehsan, M.N.; Riza, M.; Pervez, M.N.; Khyum, M.M.O.; Liang, Y.; Naddeo, V. Environmental and Health Impacts of PFAS: Sources, Distribution and Sustainable Management in North Carolina (USA). Sci. Total Environ. 2023, 878, 163123. [Google Scholar] [CrossRef]
- Li, Y.-J.; Yuan, Y.; Tan, W.-B.; Xi, B.-D.; Wang, H.; Hui, K.-L.; Chen, J.-B.; Zhang, Y.-F.; Wang, L.-F.; Li, R.-F. Antibiotic Resistance Genes and Heavy Metals in Landfill: A Review. J. Hazard. Mater. 2023, 2023, 132395. [Google Scholar] [CrossRef]
- Vlaanderen, E.J.; Ghaly, T.M.; Moore, L.R.; Focardi, A.; Paulsen, I.T.; Tetu, S.G. Plastic Leachate Exposure Drives Antibiotic Resistance and Virulence in Marine Bacterial Communities. bioRxiv 2023, 15, 121558. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.Q.; Vu, H.P.; Nguyen, L.N.; Wang, Q.; Djordjevic, S.P.; Donner, E.; Yin, H.; Nghiem, L.D. Monitoring Antibiotic Resistance Genes in Wastewater Treatment: Current Strategies and Future Challenges. Sci. Total Environ. 2021, 783, 146964. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yang, Z.H.; Zheng, Y.; Wang, Q.P.; Bai, Y.; Liu, J.B.; Zhang, Y.R.; Xiong, W.P.; Lu, Y.; Fan, C.Z. Metagenomic Analysis Reveals the Effects of Long-Term Antibiotic Pressure on Sludge Anaerobic Digestion and Antimicrobial Resistance Risk. Bioresour. Technol. 2019, 282, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Vestel, J.; Caldwell, D.J.; Tell, J.; Constantine, L.; Häner, A.; Hellstern, J.; Journel, R.; Ryan, J.J.; Swenson, T.; Xei, W. Default Predicted No-Effect Target Concentrations for Antibiotics in the Absence of Data for the Protection against Antibiotic Resistance and Environmental Toxicity. Integr. Environ. Assess. Manag. 2022, 18, 863–867. [Google Scholar] [CrossRef]
- Brunn, H.; Arnold, G.; Körner, W.; Rippen, G.; Steinhäuser, K.G.; Valentin, I. PFAS: Forever Chemicals—Persistent, Bioaccumulative and Mobile. Reviewing the Status and the Need for Their Phase out and Remediation of Contaminated Sites. Environ. Sci. Eur. 2023, 35, 20. [Google Scholar] [CrossRef]
- Backhaus, T. Commentary on the EU Commission’s Proposal for Amending the Water Framework Directive, the Groundwater Directive, and the Directive on Environmental Quality Standards. Environ. Sci. Eur. 2023, 35, 22. [Google Scholar] [CrossRef]
- Thompson, K.A.; Mortazavian, S.; Gonzalez, D.J.; Bott, C.; Hooper, J.; Schaefer, C.E.; Dickenson, E.R.V. Poly- and Perfluoroalkyl Substances in Municipal Wastewater Treatment Plants in the United States: Seasonal Patterns and Meta-Analysis of Long-Term Trends and Average Concentrations. ACS Environ. Sci. Technol. Water 2022, 2, 690–700. [Google Scholar] [CrossRef]
- Tiwari, A.; Kurittu, P.; Al-Mustapha, A.I.; Heljanko, V.; Johansson, V.; Thakali, O.; Mishra, S.K.; Lehto, K.M.; Lipponen, A.; Oikarinen, S.; et al. Wastewater Surveillance of Antibiotic-Resistant Bacterial Pathogens: A Systematic Review. Front. Microbiol. 2022, 13, 977106. [Google Scholar] [CrossRef]
- Chique, C.; Cullinan, J.; Hooban, B.; Morris, D. Mapping and Analysing Potential Sources and Transmission Routes of Antimicrobial Resistant Organisms in the Environment Using Geographic Information Systems—An Exploratory Study. Antibiotics 2019, 8, 16. [Google Scholar] [CrossRef]
- Lin, X.; Yang, S.; Gong, Z.; Ni, R.; Shi, X.; Song, L. Viral Community in Landfill Leachate: Occurrence, Bacterial Hosts, Mediation Antibiotic Resistance Gene Dissemination, and Function in Municipal Solid Waste Decomposition. Sci. Total Environ. 2022, 853, 158561. [Google Scholar] [CrossRef]
- Garcete, L.A.A.; Martinez, J.E.R.; Barrera, D.B.V.; Bonugli-Santos, R.C.; Passarini, M.R.Z. Biotechnological Potential of Microorganisms from Landfill Leachate: Isolation, Antibiotic Resistance and Leachate Discoloration. Anal. Acad. Bras. Cienc. 2022, 94, e20210642. [Google Scholar] [CrossRef] [PubMed]
- Masoner, J.R.; Kolpin, D.W.; Furlong, E.T.; Cozzarelli, I.M.; Gray, J.L. Landfill Leachate as a Mirror of Today’s Disposable Society: Pharmaceuticals and Other Contaminants of Emerging Concern in Final Leachate from Landfills in the Conterminous United States. Environ. Toxicol. Chem. 2016, 35, 906–918. [Google Scholar] [CrossRef]
- Masoner, J.R.; Kolpin, D.W.; Cozzarelli, I.M.; Smalling, K.L.; Bolyard, S.C.; Field, J.A.; Furlong, E.T.; Gray, J.L.; Lozinski, D.; Reinhart, D.; et al. Landfill Leachate Contributes Per-/Poly-Fluoroalkyl Substances (PFAS) and Pharmaceuticals to Municipal Wastewater. Environ. Sci. 2020, 6, 1300–1311. [Google Scholar] [CrossRef]
- Pisharody, L.; Gopinath, A.; Malhotra, M.; Nidheesh, P.V.; Kumar, M.S. Occurrence of Organic Micropollutants in Municipal Landfill Leachate and Its Effective Treatment by Advanced Oxidation Processes. Chemosphere 2022, 287, 132216. [Google Scholar] [CrossRef]
- Rezaei Adaryani, A.; Keen, O. Occurrence of Pharmaceuticals and Plasticizers in Leachate from Municipal Landfills of Different Age. Waste Manag. 2022, 141, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, R.; Lei, Y.; Song, L. Antibiotic Resistance Genes in Landfill Leachates from Seven Municipal Solid Waste Landfills: Seasonal Variations, Hosts, and Risk Assessment. Sci. Total Environ. 2022, 853, 158677. [Google Scholar] [CrossRef]
- Zhao, R.; Liu, J.; Feng, J.; Li, X.; Li, B. Microbial Community Composition and Metabolic Functions in Landfill Leachate from Different Landfills of China. Sci. Total Environ. 2021, 767, 144861. [Google Scholar] [CrossRef] [PubMed]
- Zacs, D.; Bartkevics, V. Trace Determination of Perfluorooctane Sulfonate and Perfluorooctanoic Acid in Environmental Samples (Surface Water, Wastewater, Biota, Sediments, and Sewage Sludge) Using Liquid Chromatography—Orbitrap Mass Spectrometry. J. Chromatogr. A 2016, 1473, 109–121. [Google Scholar] [CrossRef]
- Pugajeva, I.; Ikkere, L.E.; Jansons, M.; Perkons, I.; Sukajeva, V.; Bartkevics, V. Two-Dimensional Liquid Chromatography—Mass Spectrometry as an Effective Tool for Assessing a Wide Range of Pharmaceuticals and Biomarkers in Wastewater-Based Epidemiology Studies. J. Pharm. Biomed. Anal. 2021, 205, 114–295. [Google Scholar] [CrossRef] [PubMed]
- Gabor, E.M.; De Vries, E.J.; Janssen, D.B. Efficient Recovery of Environmental DNA for Expression Cloning by Indirect Extraction Methods. FEMS Microbiol. Ecol. 2003, 44, 153–163. [Google Scholar] [CrossRef]
- Sala, M.M.; Pinhassi, J.; Gasol, J.M. Estimation of Bacterial Use of Dissolved Organic Nitrogen Compounds in Aquatic Ecosystems Using Biolog Plates. Aquat. Microb. Ecol. 2006, 42, 1–5. [Google Scholar] [CrossRef]
- Urakawa, H.; Ali, J.; Ketover, R.D.J.; Talmage, S.D.; Garcia, J.C.; Campbell, I.S.; Loh, A.N.; Parsons, M.L. Shifts of Bacterioplankton Metabolic Profiles along the Salinity Gradient in a Subtropical Estuary. Oceanography 2013, 12, 410814. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Dale, R.K.; Pedersen, B.S.; Quinlan, A.R. Pybedtools: A Flexible Python Library for Manipulating Genomic Datasets and Annotations. Bioinformatics 2011, 27, 3423–3424. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive Analysis of Metagenomics Data for Microbiome Studies and Pathogen Identification. Bioinformatics 2020, 36, 1303–1304. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of Metagenome Assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed 2017. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 21 September 2023).
- Mukherjee, S.; Mukhopadhyay, S.; Hashim, M.A.; Gupta, B. Sen Contemporary Environmental Issues of Landfill Leachate: Assessment and Remedies. Crit. Rev. Environ. Sci. Technol. 2015, 45, 472–590. [Google Scholar] [CrossRef]
- Wijekoon, P.; Koliyabandara, P.A.; Cooray, A.T.; Lam, S.S.; Athapattu, B.C.L.; Vithanage, M. Progress and Prospects in Mitigation of Landfill Leachate Pollution: Risk, Pollution Potential, Treatment and Challenges. J. Hazard. Mater. 2022, 421, 126627. [Google Scholar] [CrossRef]
- Wu, D.; Wang, B.H.; Xie, B. Validated Predictive Modelling of Sulfonamide and Beta-Lactam Resistance Genes in Landfill Leachates. J. Environ. Manag. 2019, 241, 374. [Google Scholar] [CrossRef]
- EPA. Technical Fact Sheet: Drinking Water Health Advisories for Four PFAS (PFOA, PFOS, GenX Chemicals, and PFBS). 2022. Available online: https://www.epa.gov/system/files/documents/2022-06/technical-factsheet-four-PFAS.pdf (accessed on 21 September 2023).
- Zhang, H.; Chen, Y.; Liu, Y.; Bowden, J.A.; Tolaymat, T.M.; Townsend, T.G.; Solo-Gabriele, H.M. Relationships between Per- and Polyfluoroalkyl Substances (PFAS) and Physical-Chemical Parameters in Aqueous Landfill Samples. Chemosphere 2023, 329, 138541. [Google Scholar] [CrossRef] [PubMed]
- Lachassagne, D.; Soubrand, M.; Casellas, M.; Gonzalez-Ospina, A.; Dagot, C. Impact of Sludge Stabilization Processes and Sludge Origin (Urban or Hospital) on the Mobility of Pharmaceutical Compounds Following Sludge Landspreading in Laboratory Soil-Column Experiments. Environ. Sci. Pollut. Res. 2015, 22, 17135–17150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.; Zhang, S.; Li, J.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-Antibiotic Pharmaceuticals Promote the Transmission of Multidrug Resistance Plasmids through Intra- and Intergenera Conjugation. ISME J. 2021, 15, 2493–2508. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Xia, M.; Tao, J.; Pang, Y.; Yu, F.; Wu, J.; Chen, S. Metagenomics Analysis Reveals the Microbial Communities, Antimicrobial Resistance Gene Diversity and Potential Pathogen Transmission Risk of Two Different Landfills in China. Diversity 2021, 13, 230. [Google Scholar] [CrossRef]
- Wen, P.; Huang, Y.; Qiu, Z.; Li, Q. Microbial Response during Treatment of Different Types of Landfill Leachate in a Semi-Aerobic Aged Refuse Biofilter. Chemosphere 2021, 262, 127822. [Google Scholar] [CrossRef] [PubMed]
- Saxena, V.; Padhi, S.K.; Pattanaik, L.; Bhatt, R. Simultaneous Removal of Carbon, Nitrogen, and Phosphorus from Landfill Leachate Using an Aerobic Granular Reactor. Environ. Technol. Innov. 2022, 28, 102657. [Google Scholar] [CrossRef]
- González-Cortés, J.J.; Valle, A.; Ramírez, M.; Cantero, D. Characterization of Bacterial and Archaeal Communities by DGGE and Next Generation Sequencing (NGS) of Nitrification Bioreactors Using Two Different Intermediate Landfill Leachates as Ammonium Substrate. Waste Biomass Valorization 2022, 13, 753–766. [Google Scholar] [CrossRef]
- Fan, X.T.; Li, H.; Chen, Q.L.; Zhang, Y.S.; Ye, J.; Zhu, Y.G.; Su, J.Q. Fate of Antibiotic Resistant Pseudomonas Putida and Broad Host Range Plasmid in Natural Soil Microcosms. Front. Microbiol. 2019, 10, 194. [Google Scholar] [CrossRef] [PubMed]
- Subirats, J.; Sànchez-Melsió, A.; Borrego, C.M.; Balcázar, J.L.; Simonet, P. Metagenomic Analysis Reveals That Bacteriophages Are Reservoirs of Antibiotic Resistance Genes. Int. J. Antimicrob. Agents 2016, 48, 163–167. [Google Scholar] [CrossRef]
- Dridi, B.; Fardeau, M.L.; Ollivier, B.; Raoult, D.; Drancourt, M. The Antimicrobial Resistance Pattern of Cultured Human Methanogens Reflects the Unique Phylogenetic Position of Archaea. J. Antimicrob. Chemother. 2011, 66, 2038–2044. [Google Scholar] [CrossRef] [PubMed]
- Wolak, I.; Czatzkowska, M.; Harnisz, M.; Jastrzębski, J.P.; Paukszto, Ł.; Rusanowska, P.; Felis, E.; Korzeniewska, E. Metagenomic Analysis of the Long-Term Synergistic Effects of Antibiotics on the Anaerobic Digestion of Cattle Manure. Energies 2022, 15, 1920. [Google Scholar] [CrossRef]
- Chen, Q.L.; Li, H.; Zhou, X.Y.; Zhao, Y.; Su, J.Q.; Zhang, X.; Huang, F.Y. An Underappreciated Hotspot of Antibiotic Resistance: The Groundwater near the Municipal Solid Waste Landfill. Sci. Total Environ. 2017, 609, 966–973. [Google Scholar] [CrossRef]
- Gwenzi, W.; Musiyiwa, K.; Mangori, L. Sources, Behaviour and Health Risks of Antimicrobial Resistance Genes in Wastewaters: A Hotspot Reservoir. J. Environ. Chem. Eng. 2020, 8, 102220. [Google Scholar] [CrossRef]
- Fuhrmeister, E.R.; Harvey, A.P.; Nadimpalli, M.L.; Gallandat, K.; Ambelu, A.; Arnold, B.F.; Brown, J.; Cumming, O.; Earl, A.M.; Kang, G.; et al. Evaluating the Relationship between Community Water and Sanitation Access and the Global Burden of Antibiotic Resistance: An Ecological Study. Lancet Microbe 2023, 4, e591–e600. [Google Scholar] [CrossRef]
- European Commission. Commission Implementing Decision (EU) 2022/1307. Off. J. Eur. Union 2022, L197, 117–121. [Google Scholar]




| Parameter | MDS | YDS | Pond |
|---|---|---|---|
| pH | 8.15 | 8.16 | 8.15 |
| EC (mS/cm) | 28.2 | 27.0 | 26.9 |
| N (mg/L) | 2620 | 2560 | 2620 |
| N-NH4 (mg/L) | 2400 | 1674 | 2238 |
| COD (mg/L) | 7880 | 5015 | 6425 |
| Antibiotic Class | Compounds Belonging to the Class of Antibiotics | AMR Gene | Resistance Mechanism | Presence of AMR Gene | ||
|---|---|---|---|---|---|---|
| MDS | YDS | Pond | ||||
| Lincosamides | Lincomycin and clindamycin | Erm(35) | antibiotic target alteration | - | X | X |
| Erm(42) | - | X | - | |||
| Erm(47) | X | X | X | |||
| ErmB | X | X | X | |||
| ErmC | X | - | X | |||
| ErmF | X | X | X | |||
| ErmG | X | X | X | |||
| linG | antibiotic inactivation | X | X | X | ||
| lnuB | X | - | - | |||
| lnuC | X | X | X | |||
| lnuF | - | X | X | |||
| lnuG | - | X | X | |||
| lsaE | antibiotic target protection | - | - | X | ||
| Sulfonamides | Sulfisomidin, sulfathiazole, sulfapyridine, sulfamerazine, sulfadimidine, sulfadiazine, and phthalylsulfathiazole | sul1 | antibiotic target replacement | X | X | X |
| sul2 | X | X | X | |||
| sul4 | - | X | X | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blumfelde, M.; Gudrā, D.; Začs, D.; Vonda, K.; Žorža, L.; Selga, T.; Grīnbergs, A.; Dēliņa, A.; Bartkevičs, V.; Fridmanis, D.; et al. Risks of Antibiotic Resistance Dissemination by Leachates from Municipal Landfills of Different Ages. Water 2023, 15, 3349. https://doi.org/10.3390/w15193349
Blumfelde M, Gudrā D, Začs D, Vonda K, Žorža L, Selga T, Grīnbergs A, Dēliņa A, Bartkevičs V, Fridmanis D, et al. Risks of Antibiotic Resistance Dissemination by Leachates from Municipal Landfills of Different Ages. Water. 2023; 15(19):3349. https://doi.org/10.3390/w15193349
Chicago/Turabian StyleBlumfelde, Māra, Dita Gudrā, Dzintars Začs, Kārlis Vonda, Laura Žorža, Tūrs Selga, Andrejs Grīnbergs, Aija Dēliņa, Vadims Bartkevičs, Dāvids Fridmanis, and et al. 2023. "Risks of Antibiotic Resistance Dissemination by Leachates from Municipal Landfills of Different Ages" Water 15, no. 19: 3349. https://doi.org/10.3390/w15193349
APA StyleBlumfelde, M., Gudrā, D., Začs, D., Vonda, K., Žorža, L., Selga, T., Grīnbergs, A., Dēliņa, A., Bartkevičs, V., Fridmanis, D., & Muter, O. (2023). Risks of Antibiotic Resistance Dissemination by Leachates from Municipal Landfills of Different Ages. Water, 15(19), 3349. https://doi.org/10.3390/w15193349








