Implications of Bacterial Adaptation to Phenol Degradation under Suboptimal Culture Conditions Involving Stenotrophomonas maltophilia KB2 and Pseudomonas moorei KB4
Abstract
:1. Introduction
2. Materials and Methods
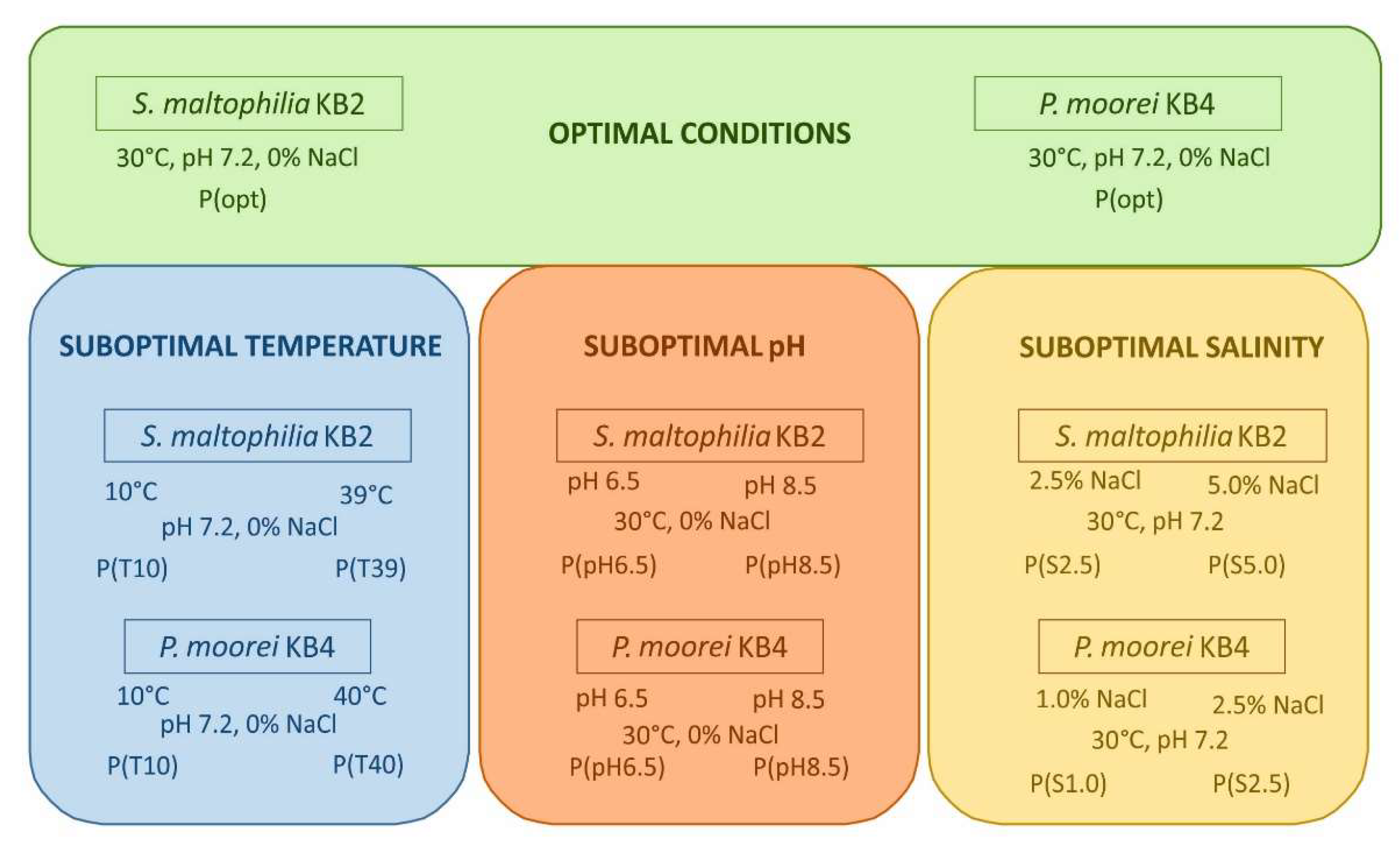
2.1. Bacterial Strains and Experimental Conditions
2.2. The Phenol Concentration Determination
2.3. The Catechol Dioxygenase Activity Determination
2.4. Determining the Whole-Cell Derived Fatty Acid Composition
2.5. Measuring the Membrane Permeability
2.6. Data Analysis
3. Results
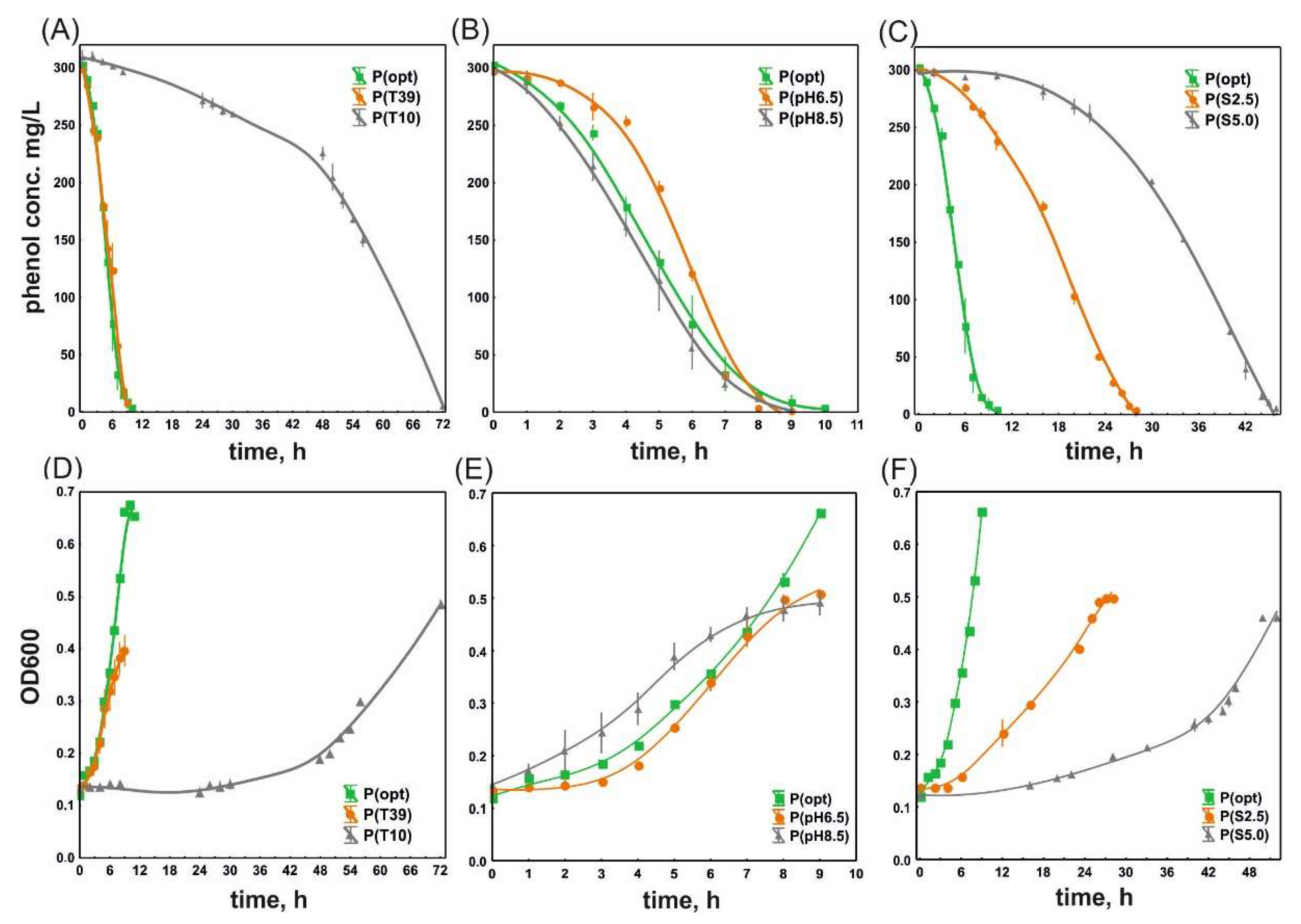
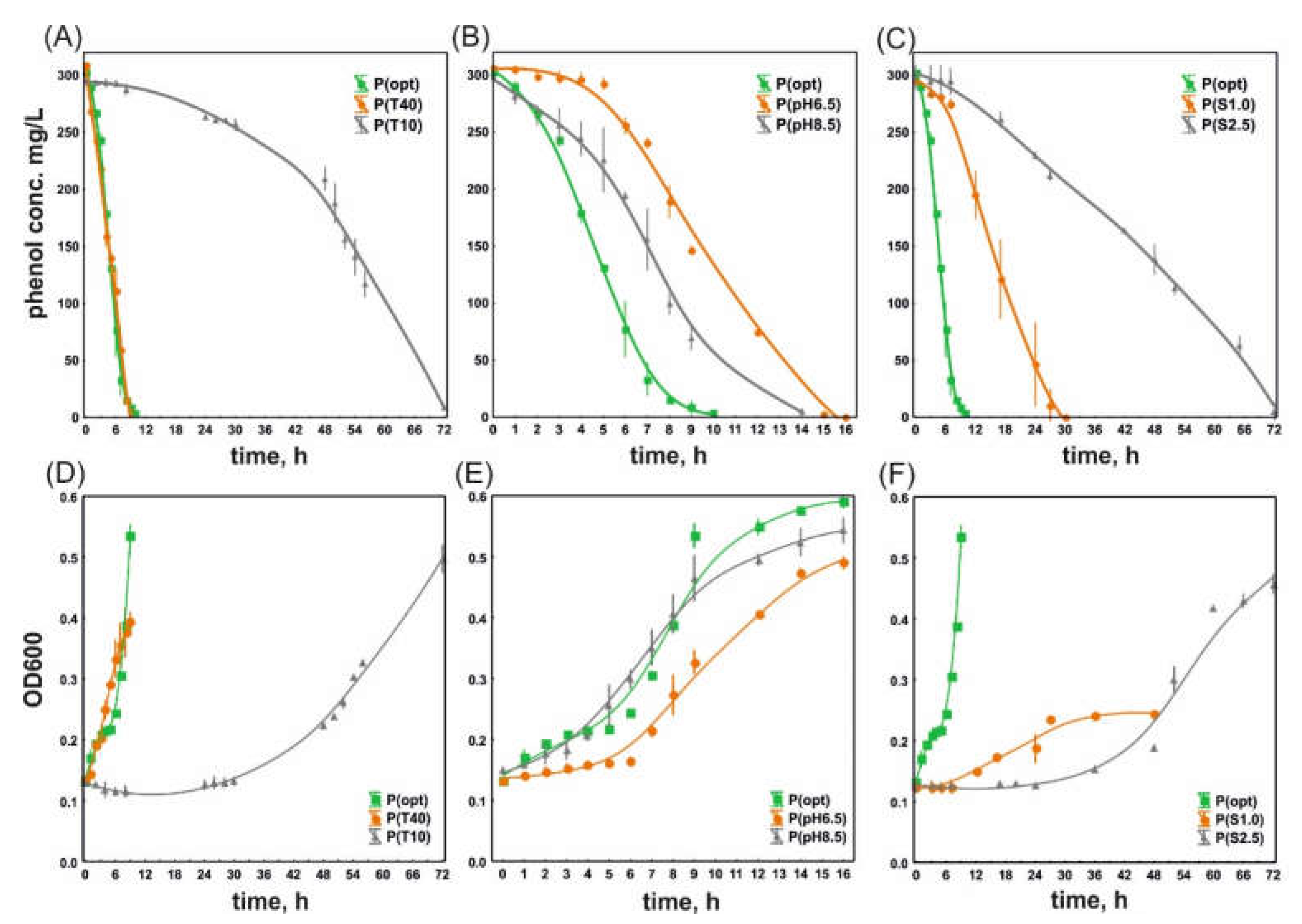
3.1. Phenol Degradation and the Growth of S. maltophilia KB2 and P. moorei KB4 under Optimal and Suboptimal Culture Conditions
3.2. The Activity of Catechol 1,2- and 2,3-dioxygenase in Bacterial Cells under Different Temperatures, pH, and Salinity
3.3. Changes in FAME Profiles of Bacteria under Various Growth Conditions
3.4. Membrane Permeability of Bacteria under Various Culture Conditions
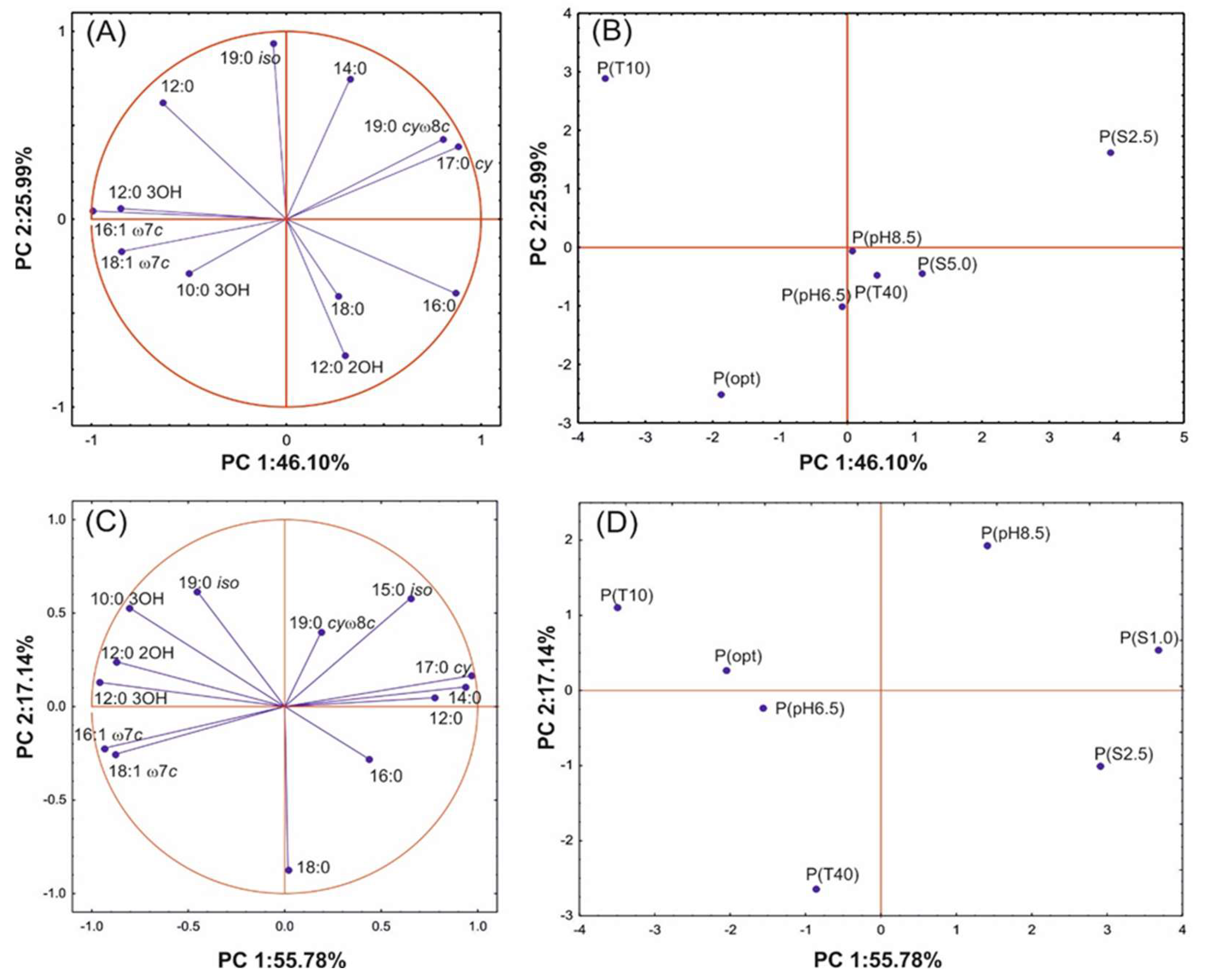
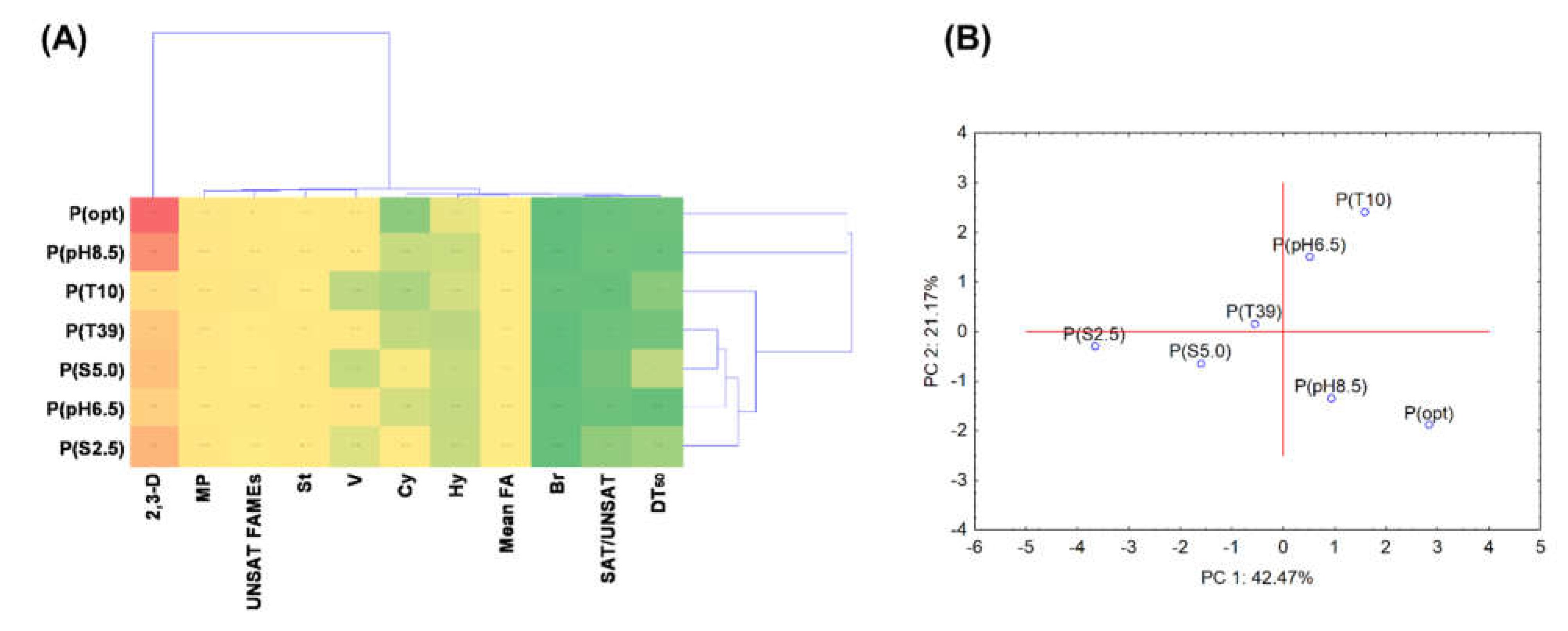
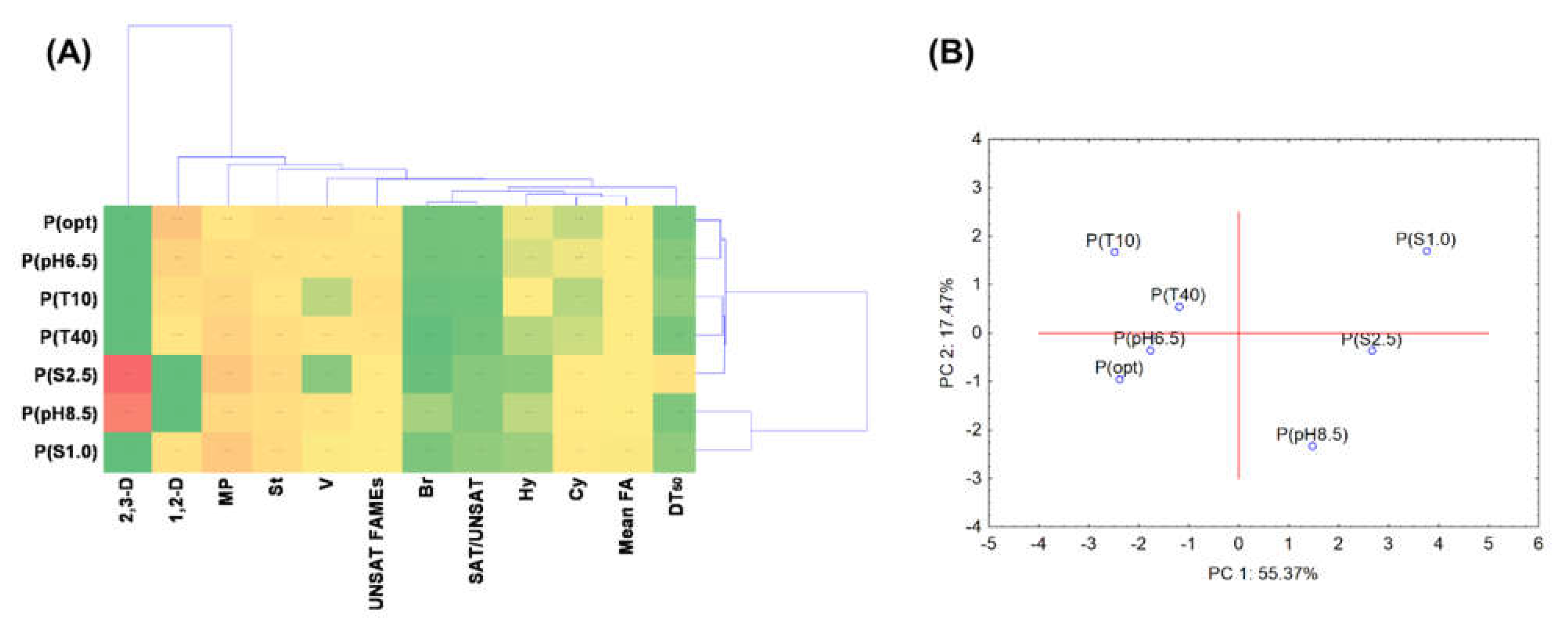
3.5. Statistical Data Exploration
4. Discussion
4.1. The Potential of S. maltophilia KB2 and P. moorei KB4 Towards Phenol Degradation under Suboptimal Conditions
4.2. The Effect of Different Temperatures, pH, and Salinity on the Membranes of S. maltophilia KB2 and P. moorei KB4 during Phenol Degradation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lochab, B.; Shuklaa, S.; Varmab, I.K. Naturally occurring phenolic sources: Monomers and polymers. RSC Adv. 2014, 4, 21712–21752. [Google Scholar] [CrossRef]
- Panigrahy, N.; Priyadarshini, A.; Sahoo, M.M.; Verma, A.K.; Daverey, A.; Sahoo, N.K. A comprehensive review on eco-toxicity and biodegradation of phenolics: Recent progress and future outlook. Environ. Technol. Innov. 2022, 27, 102423. [Google Scholar] [CrossRef]
- New Jersey Department of Health. Hazardous Substance Fact Sheet. 2015. Available online: https://nj.gov/health/eoh/rtkweb/documents/fs/1487.pdf (accessed on 12 July 2022).
- Kurnik, K.; Treder, K.; Skorupa-Kłaput, M.; Tretyn, A.; Tyburski, J. Removal of phenol from synthetic and industrial wastewater by potato pulp peroxidases. Water Air Soil Pollut. 2015, 226, 254. [Google Scholar] [CrossRef] [PubMed]
- Image Global Phenol Market-Industry Trends and Forecast to 2029. Available online: https://www.databridgemarketresearch.com/reports/global-phenol-market (accessed on 18 July 2022).
- Market Volume of Phenols Worldwide from 2015 to 2021, with a Forecast for 2022 to 2029. Available online: https://www.statista.com/statistics/979265/global-phenol-market-volume (accessed on 18 July 2022).
- Campos, F.M.; Couto, J.A.; Figueiredo, A.R.; Tóth, I.V.; Rangel, A.O.; Hogg, T.A. Cell membrane damage induced by phenolic acids on wine lactic acid bacteria. Int. J. Food Microbiol. 2009, 135, 144–151. [Google Scholar] [CrossRef]
- Muntathir, A.; Sagheer, A.; Onaizi, A. Review on phenolic wastewater remediation using homogeneous and heterogeneous enzymatic processes: Current status and potential challenges. Sep. Purif. Technol. 2019, 219, 186–207. [Google Scholar] [CrossRef]
- Downs, J.W.; Wills, B.K. Phenol toxicity. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK542311/ (accessed on 12 July 2022).
- Poi, G.; Aburto-Medina, A.; Mok, P.C.; Ball, A.S.; Shahsavari, E. Bioremediation of phenol-contaminated industrial wastewater using a bacterial consortium—From laboratory to field. Water Air Soil Pollut. 2017, 228, 89. [Google Scholar] [CrossRef]
- Duraisamy, P.; Sekar, J.; Arunkumar, A.D.; Ramalingam, P.V. Kinetics of phenol biodegradation by heavy metal tolerant rhizobacteria Glutamicibacter nicotianae MSSRFPD35 from distillery effluent contaminated soils. Front. Microbiol. 2020, 15, 1573. [Google Scholar] [CrossRef]
- Zhuang, H.; Fang, F. Bioaugmentation with phenol-degrading bacteria (PDB) as a strategy for improving start-up and stability of sequencing biofilm batch reactor (SBBR) for coal gasification wastewater (CGW) treatment. Pol. J. Environ. Stud. 2020, 29, 3955–3964. [Google Scholar] [CrossRef]
- Aljbour, S.H.; Khleifat, K.M.; Al Tarawneh, A.; Asasfeh, B.; Qaralleh, H.; El-Hasan, T.; Magharbeh, M.K.; Al-Limoun, M.O. Growth kinetics and toxicity of Pseudomonas fredriksbergsis grown on phenol as sole carbon source. J. Ecol. Eng. 2021, 22, 251–263. [Google Scholar] [CrossRef]
- Zhang, W.; Xia, X. Isolation, kinetics, and performance of a novel phenol degrading strain. Chem. Biochem. Eng. 2019, 33, 485–494. [Google Scholar] [CrossRef]
- Abarian, M.; Hassanshahian, M.; Esbah, A. Degradation of phenol at high concentrations using immobilisation of Pseudomonas putida P53 into sawdust entrapped in sodium-alginate beads. Water Sci. Technol. 2019, 79, 1387–1396. [Google Scholar] [CrossRef]
- Nogina, T.; Fomina, M.; Dumanskaya, T.; Zelena, L.; Khomenko, L.; Mikhalovsky, S.; Podgorskyi, V.; Gadd, G.M. A new Rhodococcus aetherivorans strain isolated from lubricant-contaminated soil as a prospective phenol-biodegrading agent. Appl. Microbiol. Biotechnol. 2020, 104, 3611–3625. [Google Scholar] [CrossRef]
- Barik, M.; Das, C.P.; Verma, A.K.; Sahoo, S.; Sahoo, N.K. Metabolic profiling of phenol biodegradation by an indigenous Rhodococcus pyridinivorans strain PDB9T N-1 isolated from paper pulp wastewater. Int. Biodeterior. Biodegrad. 2021, 158, 105168. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Pugazhendi, A.; Banu, R.J.; Ismail, I.M.; Qari, H.A. Biodegradation of phenol by a moderately halophilic bacterial consortium. Environ. Prog. Sustain. Energy 2018, 37, 1587–1593. [Google Scholar] [CrossRef]
- Samimi, M.; Moghadam, M.S. Phenol biodegradation by bacterial strain O-CH1 isolated from seashore. Glob. J. Environ. Sci. Manag. 2020, 6, 109–118. [Google Scholar] [CrossRef]
- Yang, X.; Hang, X.; Zhang, M.; Liu, X.; Yang, H. Relationship between acid tolerance and cell membrane in Bifidobacterium, revealed by comparative analysis of acid-resistant derivatives and their parental strains grown in medium with and without Tween 80. Appl. Microbiol. Biotechnol. 2015, 99, 5227–5236. [Google Scholar] [CrossRef]
- Bajerski, F.; Wagner, D.; Mangelsdorf, K. Cell membrane fatty acid composition of Chryseobacterium frigidisoli PB4T isolated from Antarctic glacier forefield soils in response to changing temperature and pH conditions. Front. Microbiol. 2017, 8, 677. [Google Scholar] [CrossRef]
- Dercová, K.; Murínová, S.; Dudásová, H.; Lászlová, K.; Horváthová, H. The adaptation mechanisms of bacteria applied in bioremediation of hydrophobic toxic environmental pollutants: How indigenous and introduced bacteria can respond to persistent organic pollutants-induced stress? In Persistent Organic Pollutants; Donyinah, S.K., Ed.; IntechOpen: London, UK, 2018; pp. 1–27. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- Maleki, F.; Khosravi, A.; Nasser, A.; Taghinejad, H.; Azizian, M. Bacterial heat shock protein activity. J. Clin. Diagn. Res. 2016, 10, BE01–BE03. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Rahn, T.; Kündzel, S.; Keller, A.; Neulinger, S.C. Osmotic adaptation and compatible solute biosynthesis of phototrophic bacteria as revealed from genome analyses. Microorganisms 2021, 9, 46. [Google Scholar] [CrossRef]
- Zhang, Y.; Gross, C.A. Cold shock response in bacteria. Annu. Rev. Gen. 2021, 55, 377–400. [Google Scholar] [CrossRef]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta 2005, 1717, 67–88. [Google Scholar] [CrossRef]
- Bremer, E.; Kramer, R. Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 2019, 73, 313–334. [Google Scholar] [CrossRef]
- Unell, M.; Kabelitz, N.; Jansson, J.K.; Heipieper, H.J. Adaptation of the psychrotroph Arthrobacter chlorophenolicus A6 to growth temperature and the presence of phenols by changes in the anteiso/iso ratio of branched fatty acids. FEMS Microbiol. Lett. 2007, 266, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.; Żur-Pińska, J.; Piński, A.; Pacek, G.; Mrozik, A. Adaptation of phenol-degrading Pseudomonas putida KB3 to suboptimal growth condition: A focus on degradative rate, membrane properties and expression of xylE and cfaB genes. Ecotoxicol. Environ. Saf. 2021, 221, 112431. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, C.C.C.R.; Fischer, M.A.; Kirsten, S.; Würz, B.; Wick, L.Y.; Heipieper, H.J. Adaptive response of Rhodococcus opacus PWD4 to salt and phenolic stress on the level of mycolic acids. AMB Express 2016, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Popa, P.; Timofti, M.; Voiculescu, M.; Dragan, S.; Trif, C.; Georgescu, L.P. Study of physico-chemical characteristics of wastewater in an urban agglomeration in Romania. Sci. World J. 2012, 2012, 549028. [Google Scholar] [CrossRef]
- Funamizu, N.; Takakuwa, T. Simulation analysis of operation conditions for a municipal wastewater treatment plant at low temperatures. In Biotechnological Applications of Cold-Adapted Organisms; Margesin, R., Schinner, F., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 203–220. [Google Scholar] [CrossRef]
- Bandara, W.M.; Kindaichi, T.; Satoh, H.; Sasakawa, M.; Nakahara, Y.; Takahashi, M.; Okabe, S. Anaerobic treatment of municipal wastewater at ambient temperature: Analysis of archaeal community structure and recovery of dissolved methane. Water Res. 2012, 46, 5756–5764. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.F.; Gilich, A. Opportunities to decarbonise heat in the UK using urban wastewater heat recovery. Build. Serv. Eng. Res. Technol. 2021, 42, 715–732. [Google Scholar] [CrossRef]
- Castillo-Carvajal, L.C.; Sanz-Martin, J.L.; Barragan-Huerta, B.E. Biodegradation of organic pollutants in saline wastewater by halophilic microorganisms: A review. Environ. Sci. Pollut. Res. 2014, 21, 9578–9588. [Google Scholar] [CrossRef]
- Zhuo, Y.; Sheng, X.; Cao, G. Treatment of high salinity wastewater using CWPO process for reuse. J. Adv. Oxid. Technol. 2017, 20, 20170024. [Google Scholar] [CrossRef]
- Guzik, U.; Greń, I.; Wojcieszyńska, D.; Łabużek, S. Isolation and characterisation of a novel strain of Stenotrophomonas maltophilia possessing various dioxygenases for monocyclic hydrocarbon degradation. Braz. J. Microbiol. 2009, 40, 285–291. [Google Scholar] [CrossRef]
- Wojcieszyńska, D.; Guzik, U.; Greń, I.; Perkosz, M.; Hupert-Kocurek, K. Induction of aromatic ring: Cleavage dioxygenases in Stenotrophomonas maltophilia strain KB2 in cometabolic systems. World J. Microbiol. Biotechnol. 2011, 27, 805–811. [Google Scholar] [CrossRef]
- Nowak, A.; Greń, I.; Mrozik, A. Changes in fatty acid composition of Stenotrophomonas maltophilia KB2 during co-metabolic degradation of monochlorophenols. World J. Microbiol. Biotechnol. 2016, 32, 198. [Google Scholar] [CrossRef]
- Żur, J.; Wojcieszyńska, D.; Hupert-Kocurek, K.; Marchlewicz, A.; Guzik, U. Paracetamol–toxicity and microbial utilization. Pseudomonas moorei KB4 as a case study for exploring degradation pathway. Chemosphere 2018, 206, 192–202. [Google Scholar] [CrossRef]
- Żur, J.; Piński, A.; Michalska, J.; Hupert-Kocurek, K.; Nowak, A.; Wojcieszyńska, D.; Guzik, U. A whole-cell immobilization system on bacterial cellulose for the paracetamol-degrading Pseudomonas moorei KB4 strain. Int. Biodeterior. Biodegrad. 2020, 149, 104919. [Google Scholar] [CrossRef]
- Mrozik, A.; Piotrowska-Seget, Z.; Łabużek, S. FAME profiles in Pseudomonas vesicularis during catechol and phenol degradation in the presence of glucose as an additional carbon source. Pol. J. Microbiol. 2007, 56, 157–164. [Google Scholar]
- Mrozik, A.; Swędzioł, Ż.; Miga, S. Comparative study of phenol degradation with a wild-type and genetically modified P. vesicularis (pBR322). Plasmid stability and FAME profiling. Environ. Prot. Eng. 2015, 41, 137–155. [Google Scholar] [CrossRef]
- Sasser, M. Microbial Identification by Gas Chromatographic Analysis of Fatty Acid Methyl Esters (GC-FAME); Technical Note #101; MIDI Inc.: Newark, DE, USA, 1990; p. 101. [Google Scholar]
- Halder, S.; Yadav, K.K.; Sarkar, R.; Mukherjee, S.; Saha, P.; Haldar, S.; Karmakar, S.; Sen, T. Alteration of zeta potential and membrane permeability in bacteria: A study with cationic agents. SpringerPlus 2015, 4, 672. [Google Scholar] [CrossRef] [Green Version]
- Vijay, P.T.A.; Sahila, M.; Kunhi, A.A.M. Soil bioaugmentation with Pseudomonas aeruginosa S-CSR-0013 eliminates the inhibitory effect of phenol on germination of chickpea (Cicer arietinum) seeds. J. Environ. Sci. Pollut. Res. 2020, 6, 420–425. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, J.; Wei, X.; Lin, W.; Jiang, L. Bioaugmentation technology for treatment of toxic and refractory organic waste water based on artificial intelligence. Front. Bioeng. Biotechnol. 2021, 9, 696166. [Google Scholar] [CrossRef]
- Michalska, J.; Piński, A.; Żur, J.; Mrozik, A. Selecting bacteria candidates for the bioaugmentation of activated sludge to improve the aerobic treatment of landfill leachate. Water 2020, 12, 140. [Google Scholar] [CrossRef]
- Loh, K.-C.; Chua, S.-S. Ortho pathway of benzoate degradation in Pseudomonas putida: Induction of meta pathway at high substrate concentrations. Enzym. Microb. Technol. 2002, 30, 620–626. [Google Scholar] [CrossRef]
- Hupert-Kocurek, K.; Guzik, U.; Wojcieszyńska, D. Characterization of catechol 2,3-dioxygenase from Planococcus sp. strain S5 induced by high phenol concentration. Acta Biochim. Pol. 2012, 59, 345–351. [Google Scholar] [CrossRef]
- Silva, A.S.; Camargo, F.A.O.; Andreazza, R.; Jacques, R.J.S.; Baldoni, D.B.; Bento, F.M. Enzymatic Activity of catechol 1,2-dioxygenase and catechol 2,3-dioxygenase produced by Gordonia polyisoprenivorans. Quím. Nova 2012, 35, 1587–1592. [Google Scholar] [CrossRef]
- Mei, R.; Zhou, M.; Xu, L.; Zhang, Y.; Su, X. Characterization of a pH-tolerant strain Cobetia sp. SASS1 and its phenol degradation performance under salinity condition. Front. Microbiol. 2019, 10, 2034. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, K.; Wang, H.; Shang, Y.; Yang, X. Characteristics of phenol degradation in saline conditions of a halophilic strain JS3 isolated from industrial activated sludge. Mar. Pollut. Bull. 2015, 99, 230–234. [Google Scholar] [CrossRef]
- Żur, J.; Piński, A.; Wojcieszyńska, D.; Smułek, W.; Guzik, U. Diclofenac degradation—Enzymes, genetic background and cellular alterations triggered in diclofenac-metabolizing strain Pseudomonas moorei KB4. Int. J. Mol. Sci. 2020, 21, 6786. [Google Scholar] [CrossRef]
- Klebensberger, J.; Rui, O.; Fritz, E.; Schink, B.; Philipp, B. Cell aggregation of Pseudomonas aeruginosa strain PAO1 as an energy-dependent stress response during growth with sodium dodecyl sulfate. Arch. Microbiol. 2006, 185, 417–427. [Google Scholar] [CrossRef] [Green Version]
- Hahn, M.W.; Moore, E.R.B.; Höfle, M.G. Role of microcolony formation in the protistan grazing defense of the aquatic bacterium Pseudomonas sp. Microb. Ecol. 2000, 39, 175–185. [Google Scholar] [CrossRef]
- Farrell, A.; Quilty, B. Substrate-dependent autoaggregation of Pseudomonas putida CP1 during the degradation of mono-chlorophenols and phenol. J. Ind. Microbiol. Biotechnol. 2002, 28, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 1–4, 140–164. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.I.; Saha, S.C.; Deplazes, E. Phenolic compounds alter the ion permeability of phospholipid bilayers via specific lipid interactions. Phys Chem Chem Phys 2021, 23, 22352–22366. [Google Scholar] [CrossRef]
- Sierra, J.D.M.; Oosterkamp, M.J.; Wang, W.; Spanjers, H.; van Lier, J.B. Impact of long-term salinity exposure in anaerobic membrane bioreactors treating phenolic wastewater: Performance robustness and endured microbial community. Water Res. 2018, 141, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Poger, D.; Mark, A.E. A ring to rule them all: The effect of cyclopropane fatty acids on the fluidity of lipid bilayers. J. Phys. Chem. B 2015, 119, 5487–5495. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Ding, L.; Xu, K.; Geng, J.; Ren, H. Effect of low temperature on highly unsaturated fatty acid biosynthesis in activated sludge. Bioresour. Technol. 2016, 211, 494–501. [Google Scholar] [CrossRef]
- Hassan, N.; Anesio, A.M.; Rafiq, M.; Holtvoeth, J.; Bull, I.; Haleem, A.; Shah, A.A.; Hasan, F. temperature driven membrane lipid adaptation in glacial psychrophilic bacteria. Front. Microbiol. 2020, 11, 824. [Google Scholar] [CrossRef]
- Wang, C.; Bendle, J.; Yang, Y.; Yang, H.; Sun, H.; Huang, J.; Xie, S. Impacts of pH and temperature on soil bacterial 3-hydroxy fatty acids: Development of novel terrestrial proxies. Org. Geochem. 2016, 94, 21–31. [Google Scholar] [CrossRef]
- Bale, N.J.; Rijpstra, W.I.C.; Sahonero-Canavesi, D.X.; Oshkin, I.Y.; Belova, S.E.; Dedysh, S.N.; Sinninghe Damsté, J.S. Fatty acid and hopanoid adaption to cold in the methanotroph Methylovulum psychrotolerans. Front. Microbiol. 2019, 10, 589. [Google Scholar] [CrossRef] [Green Version]
- Papazi, A.; Pappas, I.; Kotzabasis, K. Combinational system for biodegradation of olive oil mill wastewater phenolics and high yield of bio-hydrogen production. J. Biotechnol. 2019, 306, 47–53. [Google Scholar] [CrossRef]
| Experiment | k, /h | DT50, h | V, mg/L·h |
|---|---|---|---|
| S. maltophilia KB2 | |||
| P(opt) | 0.448 ± 0.009 a | 1.55 ± 0.03 ab | 51.23 ± 0.98 b |
| P(T10) | 0.167 ± 0.024 b | 4.21 ± 0.65 d | 9.17 ± 0.33 a |
| P(T39) | 0.352 ± 0.095 a | 2.09 ± 0.66 a | 39.39 ± 2.70 c |
| P(pH6.5) | 1.036 ± 0.104 c | 0.67 ± 0.06 c | 62.33 ± 0.83 d |
| P(pH8.5) | 0.658 ± 0.053 d | 1.06 ± 0.09 bc | 48.99 ± 1.20 e |
| P(S2.5) | 0.108 ± 0.002 b | 6.42 ± 0.10 e | 12.48 ± 0.12 f |
| P(S5.0) | 0.076 ± 0.004 b | 9.17 ± 0.52 f | 10.07 ± 0.23 a |
| P. moorei KB4 | |||
| P(opt) | 0.323 ± 0.041 a | 2.17 ± 0.26 a | 46.03 ± 8.22 a |
| P(T10) | 0.137 ± 0.011 b | 5.09 ± 0.38 b | 8.94 ± 0.51 c |
| P(T40) | 0.307 ± 0.081 a | 2.39 ± 0.74 a | 35.53 ± 2.19 b |
| P(pH6.5) | 0.190 ± 0.020 bc | 3.67 ± 0.38 c | 36.16 ± 2.81 b |
| P(pH8.5) | 0.254 ± 0.040 ac | 2.78 ± 0.48 a | 34.97 ± 4.21 b |
| P(S1.0) | 0.154 ± 0.018 b | 4.51 ± 0.49 b | 15.16 ± 0.13 d |
| P(S2.5) | 0.021 ± 0.003 d | 33.42 ± 0.96 d | 3.97 ± 0.16 e |
| Experiment | Experiment | |||
|---|---|---|---|---|
| S. maltophilia KB2 | P. moorei KB4 | |||
| 2,3-D | 1,2-D | 2,3-D | ||
| mU/mL | mU/mL | |||
| P(opt) | 769 ± 73 a | P(opt) | 77.42 ± 1.10 a | 0.0 ± 0.0 b |
| P(T10) | 85 ± 50 b | P(T10) | 6.38 ± 1.00 b | 0.0 ± 0.0 b |
| P(T39) | 282 ± 38 c | P(T40) | 4.40 ± 0.30 c | 0.0 ± 0.0 b |
| P(pH6.5) | 229 ± 29 c | P(pH6.5) | 53.90 ± 1.60 e | 0.0 ± 0.0 b |
| P(pH8.5) | 552 ± 61 e | P(pH8.5) | 0.00 ± 0.00 d | 189.2 ± 33.2 a |
| P(S2.5) | 330 ± 30 c | P(S1.0) | 29.04 ± 1.00 f | 0.0 ± 0.0 b |
| P(S5.0) | 290 ± 69 c | P(S2.5) | 0.00 ± 0.00 d | 171.2 ± 62.2 a |
| Experiment | Mean FA | SAT/UNSAT | Experiment | Mean FA | SAT/UNSAT |
|---|---|---|---|---|---|
| S. maltophilia KB2 | P. moorei KB4 | ||||
| P(opt) | 15.70 ± 0.02 a | 1.05 ± 0.04 a | P(opt) | 15.59 ± 0.12 a | 1.98 ± 0.16 a |
| P(T10) | 15.81 ± 0.04 ac | 0.77 ± 0.06 b | P(T10) | 15.43 ± 0.07 b | 1.08 ± 0.06 b |
| P(T39) | 16.03 ± 0.02 b | 1.53 ± 0.01 a | P(T40) | 16.02 ± 0.06 c | 1.55 ± 0.01 c |
| P(pH6.5) | 15.94 ± 0.04 bc | 1.35 ± 0.20 a | P(pH6.5) | 15.81 ± 0.06 d | 1.78 ± 0.30 ac |
| P(pH8.5) | 15.86 ± 0.05 b | 1.37 ± 0.00 a | P(pH8.5) | 15.78 ± 0.03 d | 3.77 ± 0.00 d |
| P(S2.5) | 15.99 ± 0.03 b | 5.01 ± 0.03 c | P(S1.0) | 15.05 ± 0.06 e | 4.79 ± 0.02 e |
| P(S5.0) | 15.81 ± 0.02 a | 2.23 ± 0.00 d | P(S2.5) | 15.59 ± 0.11 a | 3.66 ± 0.02 d |
| Experiment | MP, % | Experiment | MP, % |
|---|---|---|---|
| S. maltophilia KB2 | P. moorei KB4 | ||
| P(opt) | 52.66 ± 2.33 a | P(opt) | 28.40 ± 1.62 a |
| P(T10) | 57.10 ± 2.09 c | P(T10) | 55.03 ± 3.01 c |
| P(T39) | 60.95 ± 3.22 c | P(T40) | 68.64 ± 2.22 d |
| P(pH6.5) | 43.79 ± 2.95 b | P(pH6.5) | 42.60 ± 3.29 b |
| P(pH8.5) | 60.95 ± 1.67 c | P(pH8.5) | 53.85 ± 4.94 c |
| P(S2.5) | 53.85 ± 5.02 ab | P(S1.0) | 89.94 ± 3.14 e |
| P(S5.0) | 49.41 ± 4.60 ab | P(S2.5) | 93.49 ± 5.38 e |
| DT50 | V | 2,3-D | Mean FA | SAT/ UNSAT | St | Hy | Br | Cy | UNSAT | MP | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DT50 | 1.000 | −0.874 * | −0.302 | −0.046 | 0.551 | 0.338 | −0.158 | −0.076 | 0.566 | −0.554 | −0.164 |
| V | −0.874 * | 1.000 | 0.425 | −0.001 | −0.449 | 0.014 | 0.130 | −0.137 | −0.477 | 0.320 | −0.160 |
| 2,3-D | −0.302 | 0.425 | 1.000 | −0.528 | −0.056 | 0.125 | 0.680 | −0.662 | −0.209 | 0.020 | 0.156 |
| Mean FA | −0.046 | −0.001 | −0.528 | 1.000 | 0.466 | 0.441 | −0.870 * | 0.196 | 0.526 | −0.501 | 0.157 |
| SAT/UNSAT | 0.551 | −0.449 | -0.056 | 0.466 | 1.000 | 0.548 | −0.311 | −0.026 | 0.979 ** | −0.938 ** | −0.093 |
| St | 0.338 | 0.014 | 0.125 | 0.441 | 0.548 | 1.000 | −0.488 | −0.616 | 0.505 | −0.795 | −0.067 |
| Hy | −0.158 | 0.130 | 0.680 | −0.870 * | −0.311 | −0.488 | 1.000 | -0.084 | −0.429 | 0.438 | −0.106 |
| Br | −0.076 | −0.137 | −0.662 | 0.196 | −0.026 | −0.616 | -0.084 | 1.000 | 0.078 | 0.245 | −0.382 |
| Cy | 0.566 | −0.477 | −0.209 | 0.526 | 0.979 ** | 0.505 | −0.429 | 0.078 | 1.000 | −0.919 ** | −0.136 |
| UNSAT | −0.554 | 0.320 | 0.020 | −0.501 | −0.938 ** | −0.795 * | 0.438 | 0.245 | −0.919 ** | 1.000 | 0.150 |
| MP | −0.164 | −0.160 | 0.156 | 0.157 | −0.093 | −0.067 | −0.106 | −0.382 | −0.136 | 0.150 | 1.000 |
| DT50 | V | 1,2-D | 2,3-D | Mean FA | SAT/UNSAT | St | Hy | Br | Cy | UNSAT | MP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DT50 | 1.000 | −0.664 | −0.500 | 0.705 | −0.091 | 0.323 | 0.495 | −0.576 | −0.296 | 0.454 | −0.355 | 0.617 |
| V | −0.664 | 1.000 | 0.540 | −0.328 | 0.545 | −0.305 | −0.153 | 0.336 | 0.264 | −0.372 | 0.162 | −0.757 |
| 1,2-D | −0.500 | 0.540 | 1.000 | −0.756 * | −0.072 | −0.502 | −0.371 | 0.649 | −0.319 | −0.625 | 0.450 | −0.712 |
| 2,3-D | 0.705 | −0.328 | −0.756 * | 1.000 | 0.142 | 0.511 | 0.466 | −0.557 | 0.412 | 0.630 | −0.576 | 0.392 |
| Mean FA | −0.091 | 0.545 | −0.072 | 0.142 | 1.000 | −0.522 | −0.239 | 0.104 | −0.033 | −0.367 | 0.380 | −0.375 |
| SAT/UNSAT | 0.323 | −0.305 | −0.502 | 0.511 | −0.522 | 1.000 | 0.823 | −0.777 * | 0.504 | 0.951 ** | −0.975 ** | 0.626 |
| St | 0.495 | −0.153 | −0.371 | 0.466 | −0.239 | 0.823 * | 1.000 | −0.910 ** | 0.105 | 0.785 * | −0.875 ** | 0.645 |
| Hy | −0.576 | 0.336 | 0.649 | −0.557 | 0.104 | −0.777 * | −0.910 ** | 1.000 | −0.077 | −0.837 * | 0.794 * | −0.826 * |
| Br | −0.296 | 0.264 | −0.319 | 0.412 | −0.033 | 0.504 | 0.105 | −0.077 | 1.000 | 0.472 | −0.518 | −0.164 |
| Cy | 0.454 | −0.372 | −0.625 | 0.630 | −0.367 | 0.951 ** | 0.785 * | −0.837 * | 0.472 | 1.000 | −0.940 ** | 0.674 |
| UNSAT | −0.355 | 0.162 | 0.450 | −0.576 | 0.380 | −0.975 ** | −0.875 ** | 0.794 * | −0.518 | −0.940 ** | 1.000 | −0.529 |
| MP | 0.617 | −0.757 * | −0.712 | 0.392 | −0.375 | 0.626 | 0.645 | −0.826 * | −0.164 | 0.674 | −0.529 | 1.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak, A.; Wasilkowski, D.; Mrozik, A. Implications of Bacterial Adaptation to Phenol Degradation under Suboptimal Culture Conditions Involving Stenotrophomonas maltophilia KB2 and Pseudomonas moorei KB4. Water 2022, 14, 2845. https://doi.org/10.3390/w14182845
Nowak A, Wasilkowski D, Mrozik A. Implications of Bacterial Adaptation to Phenol Degradation under Suboptimal Culture Conditions Involving Stenotrophomonas maltophilia KB2 and Pseudomonas moorei KB4. Water. 2022; 14(18):2845. https://doi.org/10.3390/w14182845
Chicago/Turabian StyleNowak, Agnieszka, Daniel Wasilkowski, and Agnieszka Mrozik. 2022. "Implications of Bacterial Adaptation to Phenol Degradation under Suboptimal Culture Conditions Involving Stenotrophomonas maltophilia KB2 and Pseudomonas moorei KB4" Water 14, no. 18: 2845. https://doi.org/10.3390/w14182845
APA StyleNowak, A., Wasilkowski, D., & Mrozik, A. (2022). Implications of Bacterial Adaptation to Phenol Degradation under Suboptimal Culture Conditions Involving Stenotrophomonas maltophilia KB2 and Pseudomonas moorei KB4. Water, 14(18), 2845. https://doi.org/10.3390/w14182845







