MicroRNA-106a-5p Inhibited C2C12 Myogenesis via Targeting PIK3R1 and Modulating the PI3K/AKT Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Culture
2.3. Animals
2.4. Transfections and Treatment of Myoblasts and Myotubes
2.5. Real-Time Quantitative PCR
2.6. Western Blotting Analysis
2.7. Immunofluorescence Analysis
2.8. Luciferase Reporter Assays
2.9. Statistical Analyses
3. Results
3.1. The Expression Profiles of miR-106a-5p in Mice
3.2. MiR-106a-5p Suppresses Myoblast Differentiation by Inhibiting the PI3K-AKT Signaling Pathway
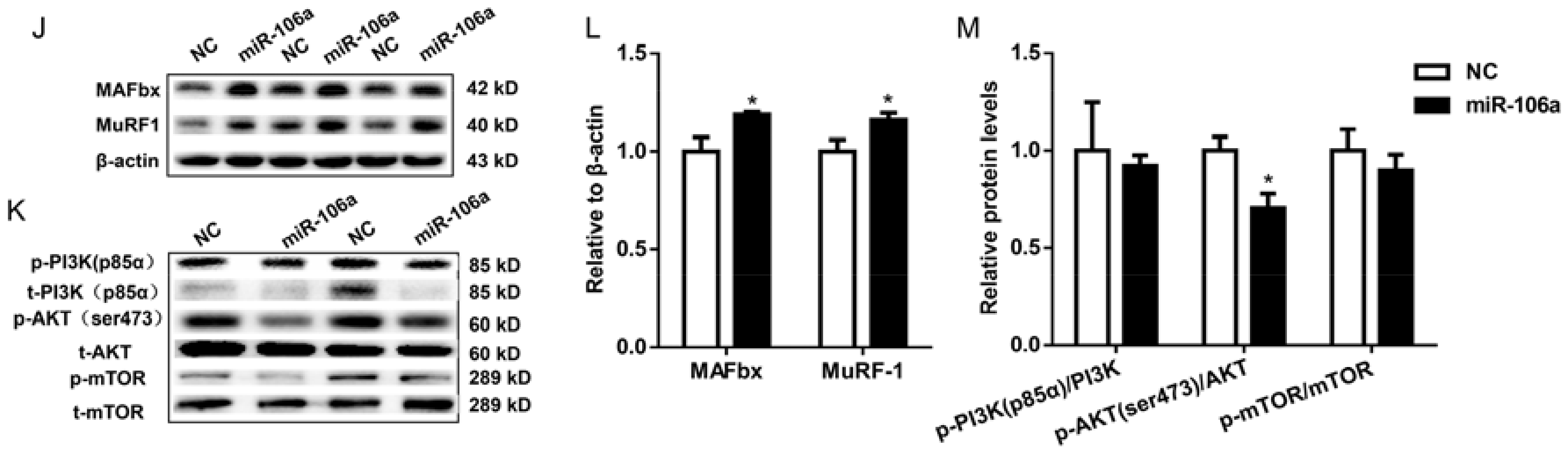
3.3. MiR-106a-5p Contributes to C2C12 Myotubes Atrophy by Suppressing PI3K-AKT Signaling Pathway
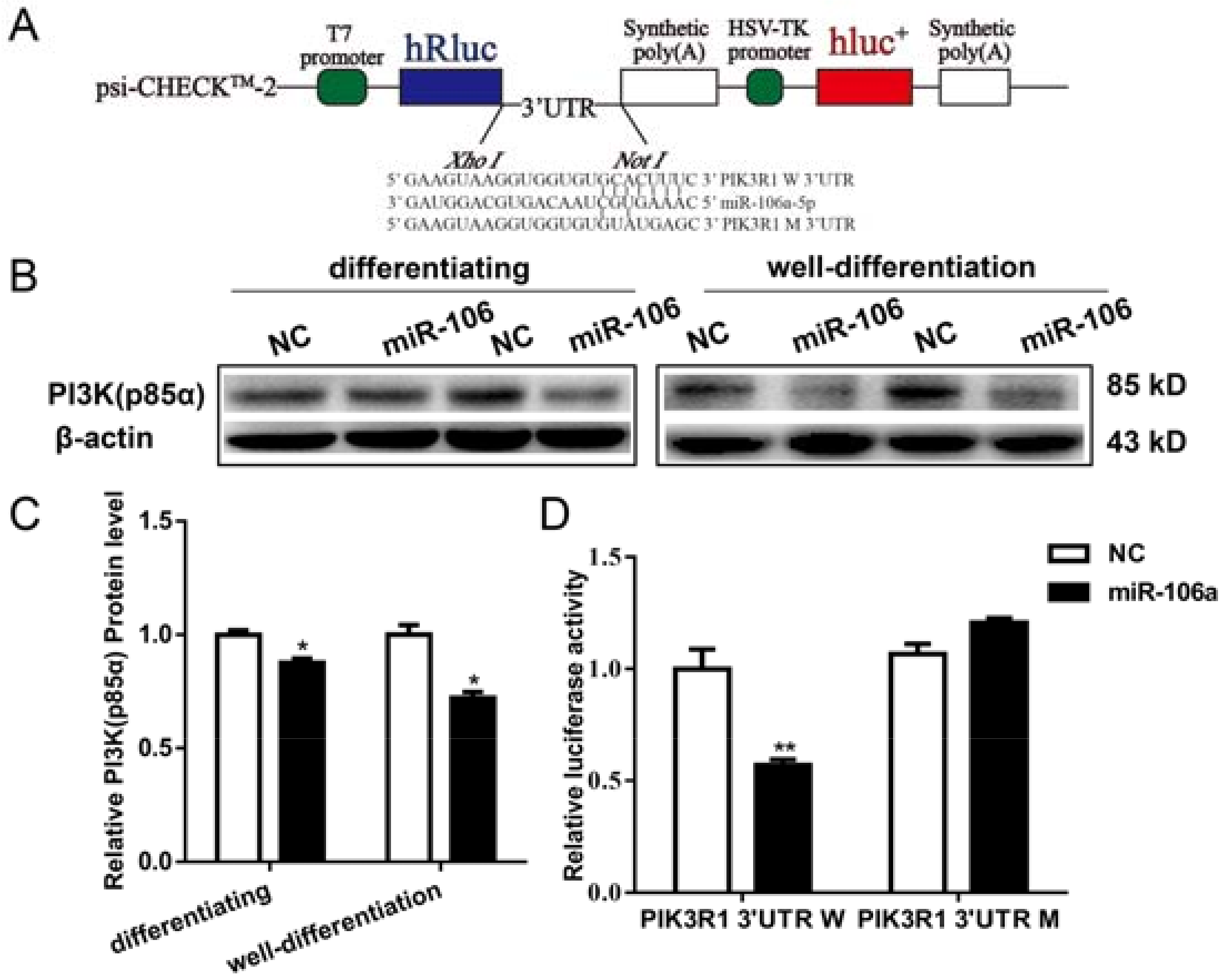
3.4. PI3K (p85α) Is a Target Gene of miR-106a-5p in Differentiating and Well-Differentiated C2C12 Cells
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cardinet, G.H. Skeletal muscle function. In Clinical Biochemistry of Domestic Animals, 5th ed.; Elsevier: Amsterdam, The Netherlands, 1997; pp. 407–440. [Google Scholar]
- Le Grand, F.; Rudnicki, M.A. Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 2007, 19, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Comai, G.; Tajbakhsh, S. Molecular and cellular regulation of skeletal myogenesis. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 110, pp. 1–73. [Google Scholar]
- Zammit, P.S. Function of the Myogenic Regulatory Factors Myf5, Myod, Myogenin and Mrf4 in Skeletal Muscle, Satellite Cells and Regenerative Myogenesis; Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Von Maltzahn, J.; Chang, N.C.; Bentzinger, C.F.; Rudnicki, M.A. Wnt signaling in myogenesis. Trends Cell Biol. 2012, 22, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, W.; Wang, L.; Liu, H.; Gong, J.; Wang, F.; Chen, X. Salidroside inhibits myogenesis by modulating p-Smad3-induced Myf5 transcription. Front. Pharmacol. 2018, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, R.; Grünhagen, J.; Becker, J.; Robinson, P.N.; Ott, C.-E.; Knaus, P. MiR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. Int. J. Biochem. Cell Biol. 2013, 45, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-N.; Baik, E.J. JAK-STAT pathway and myogenic differentiation. Jak-Stat 2013, 2, e23282. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wan, J.; Yu, B.; Diao, Y.; Zhang, W. Pip5k1α promotes myogenic differentiation via AKT activation and calcium release. Stem Cell Res. Ther. 2018, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wu, Z. The insulin-like growth factor-phosphatidylinositol 3-kinase-AKT signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J. Biol. Chem. 2000, 275, 36750–36757. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Wu, R.; Han, W.; Zhang, Y.; Zhu, D. MiR-127 enhances myogenic cell differentiation by targeting S1pPR3. Cell Death Dis. 2017, 8, e2707. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Niu, L.; Wei, W.; Zhang, W.; Li, X.; Cao, J.; Zhao, S. A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis. 2013, 4, e934. [Google Scholar] [CrossRef] [PubMed]
- Goljanek-Whysall, K.; Mok, G.; Alrefaei, A.; Kennerley, N.; Wheeler, G.; Munsterberg, A. The coordinated regulation of BAF60 variants by miR-1/206 and miR-133 clusters stabilises myogenic differentiation during embryogenesis. Int. J. Exp. Pathol. 2015, 96, A8–A9. [Google Scholar]
- Ma, M.; Wang, X.; Chen, X.; Cai, R.; Chen, F.; Dong, W.; Yang, G.; Pang, W. MicroRNA-432 targeting E2F3 and P55PIK inhibits myogenesis through PI3K/AKT/mTOR signaling pathway. RNA Biol. 2017, 14, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Jebessa, E.; Ouyang, H.; Abdalla, B.A.; Li, Z.; Abdullahi, A.Y.; Liu, Q.; Nie, Q.; Zhang, X. Characterization of miRNA and their target gene during chicken embryo skeletal muscle development. Oncotarget 2018, 9, 17309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Xu, H.; Li, R.; Wu, W.; Chao, Z.; Li, C.; Xia, W.; Wang, L.; Yang, J.; Xu, Y. Assessment of myoblast circular RNA dynamics and its correlation with miRNA during myogenic differentiation. Int. J. Biochem. Cell Biol. 2018, 99, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Khuu, C.; Jevnaker, A.-M.; Bryne, M.; Osmundsen, H. An investigation into anti-proliferative effects of microRNAs encoded by the miR-106a-363 cluster on human carcinoma cells and keratinocytes using microarray profiling of miRNA transcriptomes. Front. Genet. 2014, 5, 246. [Google Scholar] [CrossRef] [PubMed]
- Khuu, C.; Utheim, T.P.; Sehic, A. The three paralogous microRNA clusters in development and disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica 2016, 2016, 1379643. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, D.; Govindaraj, V.; Kar, S. Alteration in microRNA-17-92 dynamics accounts for differential nature of cellular proliferation. FEBS Lett. 2018, 592, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Liu, N.; Luo, L.; Zhong, J.; Tang, Z.; Kang, K.; Qu, J.; Peng, W.; Liu, L.; Li, L. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/ID1 signaling axis. Cell Death Differ. 2016, 23, 1658. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Li, G.; Yi, Z.; Nie, Q.; Zhang, X. E2f1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation. Sci. Rep. 2016, 6, 27904. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Liu, N.; Luo, L.; Kang, K.; Li, L.; Ni, R.; Qiu, H.; Gou, D. MicroRNA-17-92 regulates the transcription factor E2F3b during myogenesis in vitro and in vivo. Int. J. Mol. Sci. 2017, 18, 727. [Google Scholar] [CrossRef] [PubMed]
- Imig, J.; Brunschweiger, A.; Brümmer, A.; Guennewig, B.; Mittal, N.; Kishore, S.; Tsikrika, P.; Gerber, A.P.; Zavolan, M.; Hall, J. MiR-clip capture of a miRNA targetome uncovers a lincRNA h19–miR-106a interaction. Nat. Chem. Biol. 2015, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Mellor, P.; Furber, L.A.; Nyarko, J.N.; Anderson, D.H. Multiple roles for the p85α isoform in the regulation and function of PI3K signalling and receptor trafficking. Biochem. J. 2012, 441, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Madhala-Levy, D.; Williams, V.; Hughes, S.; Reshef, R.; Halevy, O. Cooperation between Shh and IGF-I in promoting myogenic proliferation and differentiation via the MAPK/ERK and PI3K/Akt pathways requires smo activity. J. Cell. Physiol. 2012, 227, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Mammucari, C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: Insights from genetic models. Skelet. Muscle 2011, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Briata, P.; Lin, W.-J.; Giovarelli, M.; Pasero, M.; Chou, C.-F.; Trabucchi, M.; Rosenfeld, M.G.; Chen, C.-Y.; Gherzi, R. PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death Differ. 2012, 19, 478. [Google Scholar] [CrossRef] [PubMed]
- Conejo, R.; Lorenzo, M. Insulin signaling leading to proliferation, survival, and membrane ruffling in C2C12 myoblasts. J. Cell. Physiol. 2001, 187, 96–108. [Google Scholar] [CrossRef]
- Yu, M.; Wang, H.; Xu, Y.; Yu, D.; Li, D.; Liu, X.; Du, W. Insulin-like growth factor-1 (IGF-1) promotes myoblast proliferation and skeletal muscle growth of embryonic chickens via the PI3K/Akt signalling pathway. Cell Biol. Int. 2015, 39, 910–922. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001, 3, 1009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, X.-E.; Song, C.; Sun, S.; Yang, G.; Li, X. Bambi promotes C2C12 myogenic differentiation by enhancing Wnt/β-catenin signaling. Int. J. Mol. Sci. 2015, 16, 17734–17745. [Google Scholar] [CrossRef] [PubMed]
- Foshay, K.M.; Gallicano, G.I. MiR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 2009, 326, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Glass, D.J. PI3 kinase regulation of skeletal muscle hypertrophy and atrophy. In Phosphoinositide 3-Kinase in Health and Disease; Springer: Berlin, Germany, 2010; pp. 267–278. [Google Scholar]
- Serra, C.; Palacios, D.; Mozzetta, C.; Forcales, S.V.; Morantte, I.; Ripani, M.; Jones, D.R.; Du, K.; Jhala, U.S.; Simone, C. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell 2007, 28, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Yada, T.; Ishikawa, N.; Taheruzzaman, K.; Hara, R.; Matsuzaki, T.; Nishikawa, A. Insulin-like growth factor 1 regulation of proliferation and differentiation of Xenopus laevis myogenic cells in vitro. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, I.; Eran, A.; Nishino, I.; Moggio, M.; Lamperti, C.; Amato, A.A.; Lidov, H.G.; Kang, P.B.; North, K.N.; Mitrani-Rosenbaum, S. Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA 2007, 104, 17016–17021. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, M.; Chen, M.; Zhang, K.; Gu, L.; Li, Q.; Yu, Z.; Li, N.; Meng, Q. MiR-18a induces myotubes atrophy by down-regulating IGFI. Int. J. Biochem. Cell Biol. 2017, 90, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Vogt, P.K.; Hart, J.R. Domain analysis reveals striking functional differences between the regulatory subunits of phosphatidylinositol 3-kinase (PI3K), p85α and p85β. Oncotarget 2017, 8, 55863. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Sobkiw, C.L.; Hirshman, M.F.; Logsdon, M.N.; Li, T.Q.; Goodyear, L.J.; Cantley, L.C. Loss of class IA PI3K signaling in muscle leads to impaired muscle growth, insulin response, and hyperlipidemia. Cell Metab. 2006, 3, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, N.; Alexander, M.S.; Shimizu-Motohashi, Y.; Myers, J.A.; Kawahara, G.; Kunkel, L.M. Regulation of IRS1/AKT insulin signaling by microRNA-128a during myogenesis. J. Cell Sci. 2013, 126, 2678–2691. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chan, M.C.; Yu, Y.; Bei, Y.; Chen, P.; Zhou, Q.; Cheng, L.; Chen, L.; Ziegler, O.; Rowe, G.C. MiR-29b contributes to multiple types of muscle atrophy. Nat. Commun. 2017, 8, 15201. [Google Scholar] [CrossRef] [PubMed]




| Name | Forward | Reverse |
|---|---|---|
| MyoD | CCACTCCGGGACATAGACTTG | AAAAGCGCAGGTCTGGTGAG |
| MyoG | GAGACATCCCCCTATTTCTACCA | GCTCAGTCCGCTCATAGCC |
| MyHC | GCGAATCGAGGCTCAGAACAA | GTAGTTCCGCCTTCGGTCTTG |
| Myomarker | CCTGCTGTCTCTCCCAAG | AGAACCAGTGGGTCCCTAA |
| Myomixer | GTTAGAACTGGTGAGCAGGAG | CCATCGGGAGCAATGGAA |
| MAFbx | CAGCTTCGTGAGCGACCTC | GGCAGTCGAGAAGTCCAGTC |
| MuRF1 | GTGTGAGGTGCCTACTTGCTC | GCTCAGTCTTCTGTCCTTGGA |
| β-actin | GCCATGTACGTAGCCATCCA | ACGCTCGGTCAGGATCTTCA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhu, Y.; Zhang, H.; Ma, G.; Wu, G.; Xiang, A.; Shi, X.; Yang, G.S.; Sun, S. MicroRNA-106a-5p Inhibited C2C12 Myogenesis via Targeting PIK3R1 and Modulating the PI3K/AKT Signaling. Genes 2018, 9, 333. https://doi.org/10.3390/genes9070333
Li X, Zhu Y, Zhang H, Ma G, Wu G, Xiang A, Shi X, Yang GS, Sun S. MicroRNA-106a-5p Inhibited C2C12 Myogenesis via Targeting PIK3R1 and Modulating the PI3K/AKT Signaling. Genes. 2018; 9(7):333. https://doi.org/10.3390/genes9070333
Chicago/Turabian StyleLi, Xiao, Youbo Zhu, Huifang Zhang, Guangjun Ma, Guofang Wu, Aoqi Xiang, Xin’E. Shi, Gong She Yang, and Shiduo Sun. 2018. "MicroRNA-106a-5p Inhibited C2C12 Myogenesis via Targeting PIK3R1 and Modulating the PI3K/AKT Signaling" Genes 9, no. 7: 333. https://doi.org/10.3390/genes9070333
APA StyleLi, X., Zhu, Y., Zhang, H., Ma, G., Wu, G., Xiang, A., Shi, X., Yang, G. S., & Sun, S. (2018). MicroRNA-106a-5p Inhibited C2C12 Myogenesis via Targeting PIK3R1 and Modulating the PI3K/AKT Signaling. Genes, 9(7), 333. https://doi.org/10.3390/genes9070333





