Metagenomics of Bacterial Diversity in Villa Luz Caves with Sulfur Water Springs
Abstract
1. Introduction
2. Materials and Methods
2.1. Environmental Sampling and DNA Extraction for Metagenomic Analysis
2.2. Pyrosequencing and Sequence Analysis
3. Results
3.1. Samples and Chemical Properties
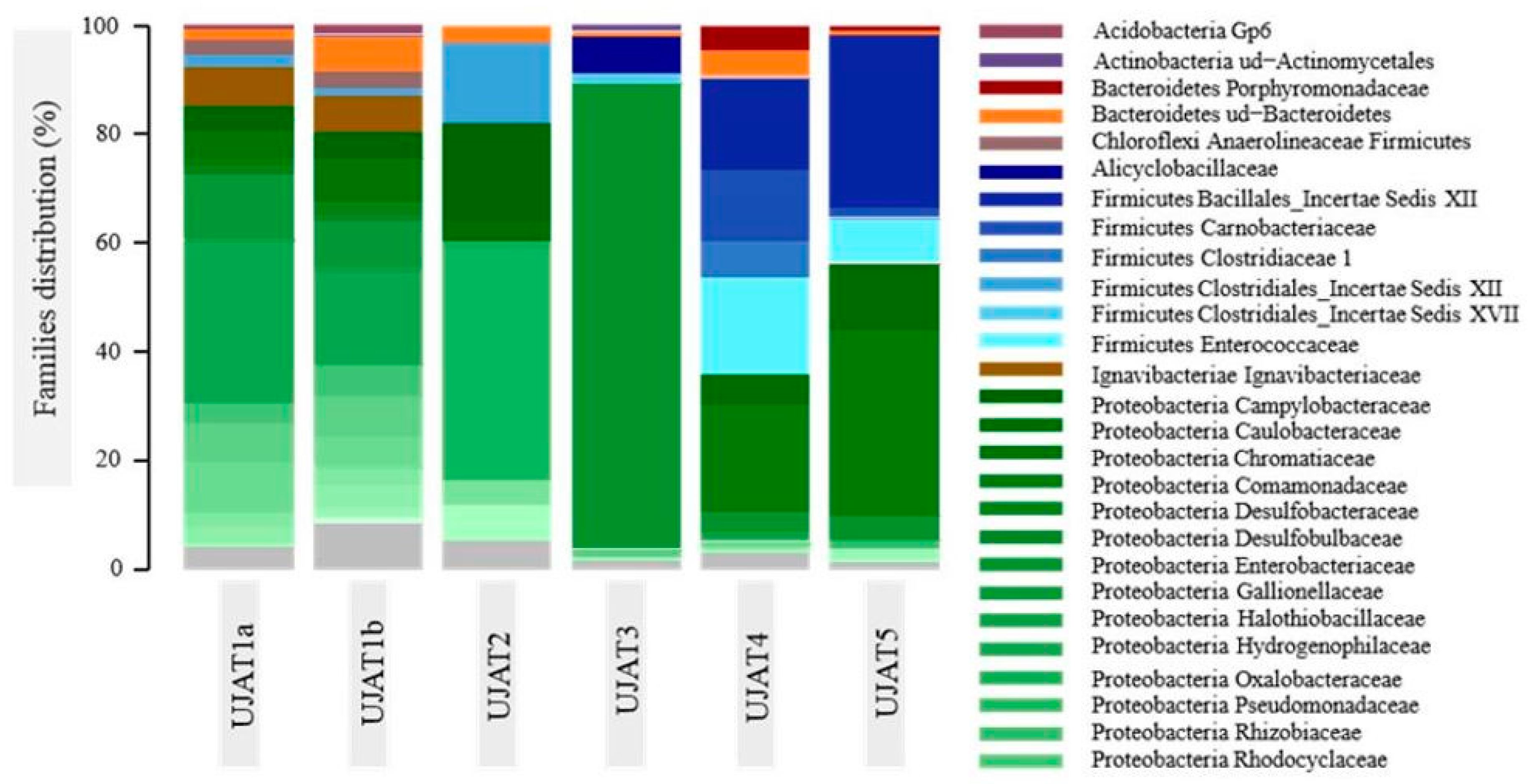
3.2. Bacterial Diversity Distribution
3.2.1. Proteobacteria
3.2.2. Firmicutes
3.2.3. Others
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Groombridge, B.; Jenkins, M.D. World Atlas of Biodiversity: Earth’s Living Resources in the 21st Century; UNEP-WCMC: Cambridge, UK; London, UK, 2002; pp. 3–30. ISBN 0-520-23668-8. [Google Scholar]
- Malhi, Y.; Grace, J. Tropical forests and atmospheric carbon dioxide. Trends Ecol. Evol. 2000, 15, 332–337. [Google Scholar] [CrossRef]
- Anantharaman, K.; Breier, J.A.; Dick, G.J. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J. 2016, 10, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Butler, R.A.; Laurance, W.F. New strategies for conserving tropical forests. Trends Ecol. Evol. 2008, 23, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Tobler, M.; Schlupp, I.; Heubel, K.U.; Riesch, R.; Garcia de León, F.J.; Giere, O.; Plath, M. Life on the edge: Hydrogen sulfide and the fish communities of a Mexican cave and surrounding waters. Extremophiles 2006, 10, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Koleff, P.; Urquiza-Haas, T.; Contreras, B. Prioridades de conservación de los bosques tropicales de méxico: Reflexiones sobre su estado de conservación y manejo. Ecosistemas 2012, 21, 6–20. [Google Scholar]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Pinar, G.; Lubitz, W.; Rolleke, S. Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol. Lett. 2002, 211, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Hose, L.D.; Palmer, A.N.; Palmer, M.V.; Northup, D.E.; Boston, P.J.; Duchene, H.R. Microbiology and geochemistry in a hydrogen-sulphide-rich karst environment. Chem. Geol. 2000, 169, 399–423. [Google Scholar] [CrossRef]
- Plath, M.; Tobler, M. Subterranean fishes of Mexico (Poecilia mexicana, Poeciliidae). In The Biology of Subterranean Fishes; Trajano, E., Bichuette, M.E., Kapoor, B.G., Eds.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2010; pp. 281–330. ISBN 978-1-57808-670-2. [Google Scholar]
- Hose, L.D.; Pisarowicz, J.A. Cueva de Villa Luz, Tabasco, Mexico: Reconnaissance Study of an Active Sulfur Spring Cave and Ecosystem. J. Cave Karst Stud. 1999, 61, 13–21. [Google Scholar]
- Tomova, I.; Lazarkevich, I.; Tomova, A.; Kambourova, M.; Vasileva-Tonkova, E. Diversity and biosynthetic potential of culturable aerobic heterotrophic bacteria isolated from Magura Cave, Bulgaria. Int. J. Speleol. 2013, 42, 65–76. [Google Scholar] [CrossRef]
- Palacios-Vargas, J.G.; Castaño-Meneses, G.; Estrada, D.A. Diversity and dynamics of microarthropods from different biotopes of Las Sardinas cave (Mexico). Subterr. Biol. 2011, 9, 113–126. [Google Scholar] [CrossRef]
- Quezada, F.; Roca, W.; Szauer, M.T.; Gómez, J.J.; López, R. Biotecnología para el uso sostenible de la biodiversidad. Capacidades Locales y Mercados Potenciales; CAF-CEPAL: Vitacura, Santiago de Chile, Chile, 2005; pp. 28–34. [Google Scholar]
- Kallmeyer, J.; Pockalny, R.; Adhikari, R.R.; Smith, D.C.; D’Hondt, S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. USA 2012, 109, 16213–16216. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.L. Identifying microorganisms responsible for ecologically significant biogeochemical processes. Nat. Rev. Microbiol. 2005, 3, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Rondon, M.R.; August, P.R.; Bettermann, A.D.; Brady, S.F.; Grossman, T.H.; Liles, M.R.; Loiacono, K.A.; Lynch, B.A.; MacNeil, I.A.; Minor, C.; et al. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000, 66, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Tang, H.; Doak, T.G.; Ye, Y. Comparing bacterial communities inferred from 16S rRNA gene sequencing and shotgun metagenomics. Pac. Symp. Biocomput. 2011, 165–176. [Google Scholar] [CrossRef]
- Engel, A.S. Microbial Diversity of Cave Ecosystems. In Geomicrobiology: Molecular and Environmental Perspective; Springer: Dordrecht, The Netherlands, 2010; pp. 219–238. [Google Scholar] [CrossRef]
- Øvreås, L.; Torsvik, V. microbial diversity and community structure in two different agricultural soil communities. Microb. Ecol. 1998, 36, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Warnes, G.R.; Bolker, B.; Gorjanc, G.; Grothendieck, G.; Korosec, A.; Lumley, T.; MacQueen, D.; Magnusson, A.; Rogers, J.; et al. GData: Various R Programming Tools for Data Manipulation. R Package Version 2.13.2. 2013. Available online: http://cran.r-project.org/package=gdata (accessed on 6 January 2017).
- DeLong, E.F.; Pace, N.R. Environmental diversity of bacteria and archaea. Syst. Biol. 2001, 50, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Hough, D.W.; Danson, M.J. Extremozymes. Curr. Opin. Chem. Biol. 2000, 3, 39–46. [Google Scholar] [CrossRef]
- Rosenberg, E.; DeLong, E.F.; Lory, S.; Stackebrandt, E.; Thompson, F. The Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Springer: Berlin/Heidelberg, Germany, 2013; pp. 39–80. [Google Scholar] [CrossRef]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam, S.A.; McGarrell, D.M.; Garrity, G.M.; Tiedje, J.M. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005, 33, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.; Barreto, C.; Kowalchuk, G.; van Veen, J.A.; Kuramae, E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [PubMed]
- Rawat, S.R.; Männistö, M.K.; Starovoytov, V.; Goodwin, L.; Hauser, M.N.L.; Land, M.; Davenport, K.W.; Woyke, T.; Häggblom, M.M. Complete genome sequence of Granulicella tundricola type strain MP5ACTX9T, an Acidobacteria from tundra soil. Stand. Genom. Sci. 2014, 9, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Lagarde, L.; Boston, P.J.; Campbell, A.R.; Hose, L.D.; Axen, G.; Stafford, K.W. Hydrogeology of northern Sierra de Chiapas, Mexico: A conceptual model based on a geochemical characterization of sulfide-rich karst brackish springs. Hydrogeol. J. 2014, 22, 1447–1467. [Google Scholar] [CrossRef]
- Simon, C.; Wiezer, A.; Strittmatter, A.W.; Daniel, R. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 2009, 75, 7519–7526. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.S.; Rosen, D.E. A cavernicolous form of the Poecilid fish Poecilia sphenops from Tabasco, Mexico. Copeia 1962, 2, 360–368. [Google Scholar] [CrossRef]
- Relman, D.A. Universal bacterial 16S rDNA amplification and sequencing. In Diagnostic Molecular Microbiology: Principles and Applications; Persing, D.H., Smith, T.F., Tenover, F.C., White, T.J., Eds.; ASM Press: Washington, DC, USA, 1993; pp. 489–495. [Google Scholar]
- Brown, M.V.; Philip, G.K.; Bunge, J.A.; Smith, M.C.; Bissett, A.; Lauro, F.M.; Fuhrman, J.A.; Donachie, S.P. Microbial community structure in the North Pacific Ocean. ISME J. 2009, 3, 1374–1386. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Huber, J.A.; Morrrison, H.G.; Sogin, M.L.; Welch, D.M. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007, 8, R143. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, B.; Taube, R.; Akyol, P.; Wemheuer, F.; Daniel, R. Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka peninsula. Archaea 2013, 2013, 136714. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 1.17-8. 2011. Available online: https://cran.r-project.org/package=vegan (accessed on 6 January 2017).
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef] [PubMed]
- Hobel, C.F.V.; Marteinsson, V.T.; Hreggvidsson, G.O.; Kristjansson, J.K. Investigation of the microbial ecology of intertidal hot springs by using diversity analysis of 16S rRNA and Chitinase genes. Appl. Environ. Microbiol. 2005, 71, 2771–2776. [Google Scholar] [CrossRef] [PubMed]
- Rampelotto, P.H. Extremophiles and Extreme Environments. Life 2013, 3, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Gonzalez, J.M.; Saiz-Jimenez, C.; Ludwig, W. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in Altamira Cave using 23S rRNA sequence analyses. Geomicrobiol. J. 2005, 22, 379–388. [Google Scholar] [CrossRef]
- Byrne, N.; Strous, M.; Crépeau, V.; Kartal, B.; Birrien, J.-L.; Schmid, M.; Lesongeur, F.; Schouten, S.; Jaeschke, A.; Jetten, M.; et al. Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J. 2009, 3, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Langner, H.W.; Donahoe-Christiansen, J.; Inskeep, W.P.; McDermott, T.R. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 2001, 3, 532–542. [Google Scholar] [CrossRef] [PubMed]
- López-López, O.; Cerdán, M.E.; González-Siso, M.I. Hot Spring Metagenomics. Life 2013, 2, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Dombard, D.R.; Shock, E.L.; Amend, J.P. Archaeal and bacterial communities in geochemically diverse hot springs of Yellowstone National Park, USA. Geobiology 2005, 3, 211–227. [Google Scholar] [CrossRef]
- Rashid, M.; Stingl, U. Contemporary molecular tools in microbial ecology and their application to advancing biotechnology. Biotechnol. Adv. 2015, 33, 1755–1773. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Ikner, L.A.; Toomey, R.S.; Nolan, G.; Neilson, J.W.; Pryor, B.M.; Maier, R.M. Culturable microbial diversity and the impact of tourism in Kartchner caverns, Arizona. Microb. Ecol. 2007, 53, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Daae, F.L.; Sandaa, R.A.; Øvreås, L. Novel techniques for analyzing microbial diversity in natural and perturbed environments. J. Biotechnol. 1998, 64, 53–62. [Google Scholar] [CrossRef]
- Johnston, M.D.; Muench, B.A.; Banks, E.D.; Barton, H.A. Human urine in Lechuguilla cave: The microbiological impact and potential for bioremediation. J. Cave Karst Stud. 2012, 74, 278–291. [Google Scholar] [CrossRef]
- Hedrich, S.; Schlömann, M.; Johnson, D.B. The iron-oxidizing proteobacteria. Microbiology 2011, 157, 1551–1564. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, E.; Nauhaus, K.; Cypionka, H.; Overmann, J. Multitude and temporal variability of ecological niches as indicated by the diversity of cultivated bacterioplankton. FEMS Microbiol. Ecol. 2001, 36, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Morse, J.L.; Berthrong, S.T.; Bernhardt, E.S.; Jackson, R.B. Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology 2007, 88, 2162–2173. [Google Scholar] [CrossRef] [PubMed]
- Catão, E.C.P.; Lopes, F.A.C.; Araújo, J.F.; de Castro, A.P.; Barreto, C.C.; Bustamante, M.M.C.; Quirino, B.F.; Krüger, R.H. Soil acidobacterial 16S rRNA gene sequences reveal subgroup level differences between Savanna-like cerrado and Atlantic forest brazilian biomes. Int. J. Microbiol. 2014, 2014, 156341:1–156341:12. [Google Scholar]
- Eichorst, S.A.; Breznak, J.A.; Schmidt, T.M. Isolation and Characterization of Soil Bacteria That Define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 2007, 73, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.; van Veen, J.A.; Kowalchuk, G.A. Comparative Analysis of Acidobacterial Genomic Fragments from Terrestrial and Aquatic Metagenomic Libraries, with Emphasis on Acidobacteria Subdivision 6. Appl. Environ. Microbiol. 2010, 76, 6769–6777. [Google Scholar] [CrossRef] [PubMed]
- Quaiser, A.; Ochsenreiter, T.; Lanz, C.; Schuster, S.C.; Treusch, A.H.; Eck, J.; Schleper, C. Acidobacteria form a coherent but highly diverse group within the bacterial domain: Evidence from environmental genomics. Mol. Microbiol. 2003, 50, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Basak, P.; Majumder, N.S.; Nag, S.; Bhattacharyya, A.; Roy, D.; Chakraborty, A.; SenGupta, S.; Roy, A.; Mukherjee, A.; Pattanayak, R.; et al. Spatiotemporal analysis of bacterial diversity in sediments of Sundarbans using parallel 16S rRNA gene Tag sequencing. Microb. Ecol. 2015, 69, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Terborgh, J. Diversity and the Tropical Rain Forest; Scientific American Library: New York, NY, USA, 1992; 242p, ISBN 0716750309. [Google Scholar]
- Crocetti, G.R.; Banfield, J.F.; Keller, J.; Bond, P.L.; Blackall, L.L. Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 2002, 148, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- LaPara, T.M.; Nakatsu, C.H.; Pantea, L.; Alleman, J.E. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl. Environ. Microbiol. 2000, 66, 3951–3959. [Google Scholar] [CrossRef] [PubMed]
- Martiny, A.C.; Albrechtsen, H.J.; Arvin, E.; Molin, S. Identification of bacteria in biofilm and bulk water samples from a nonchlorinated model drinking water distribution system: Detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl. Environ. Microbiol. 2005, 71, 8611–8617. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Pankratov, T.A.; Belova, S.E.; Kulichevskaya, I.S.; Liesack, W. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 2006, 72, 2110–2117. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, N.; Kosako, Y.; Tano, T. Acidobacterium capsulatum gen. nov., sp. nov.: An acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 1991, 22, 1–7. [Google Scholar] [CrossRef]
- Smith, S.M.; Abed, R.M.M.; Garcia-Pichel, F. Biological soil crusts of sand dunes in Cape Cod National Seashore, Massachusetts, USA. Microb. Ecol. 2004, 48, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Pinar, G.; Lubitz, W.; Rolleke, S. Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Palaeolithic paintings. Environ. Microbiol. 2002, 4, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Pinar, G.; Lubitz, W.; Rolleke, S. Phylogenetic diversity of bacteria associated with Paleolithic paintings and surrounding rock walls in two Spanish caves (Llonin and La Garma). FEMS Micrbiol. Ecol. 2004, 47, 235–247. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 2003, 14, 303–310. [Google Scholar] [CrossRef]
- Zimmermann, J.; Gonzalez, J.M.; Saiz-Jimenez, C. Epilithic biofilms in Saint Callixtus Catacombs (Rome) harbour a broad spectrum of Acidobacteria. Antonie Leeuwenhoek 2006, 89, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, W.; Bauer, S.H.; Bauer, M.; Held, I.; Kirchhof, G.; Schulze, R.; Huber, I.; Spring, S.; Hartmann, A.; Schleifer, K.H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 1997, 153, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Meisinger, D.B.; Zimmermann, J.; Ludwig, W.; Schleifer, K.H.; Wanner, G.; Schmid, M.; Bennett, P.C.; Engel, A.S.; Lee, N.M. In situ detection of novel Acidobacteria in microbial mats from a chemolithoautotrophically based cave ecosystem (Lower Kane Cave, WY, USA). Environm. Microbiol. 2007, 9, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.; Pijl, A.S.; van Veen, J.A.; Kowalchuk, G.A. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 2009, 3, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Alvarez, V.; King, G.M.; Nusslein, K. Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol. Ecol. 2007, 60, 60–73. [Google Scholar] [CrossRef] [PubMed]
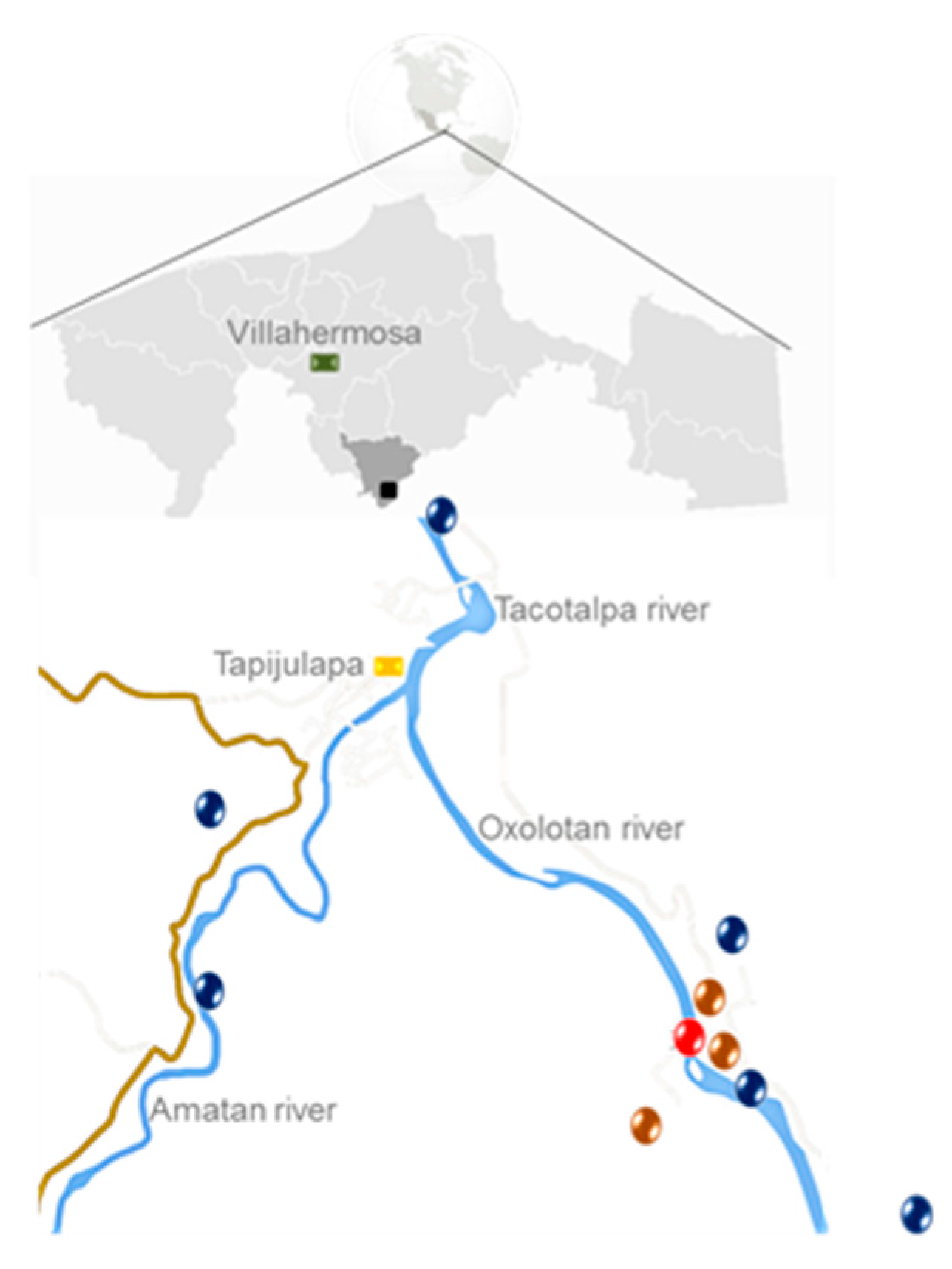
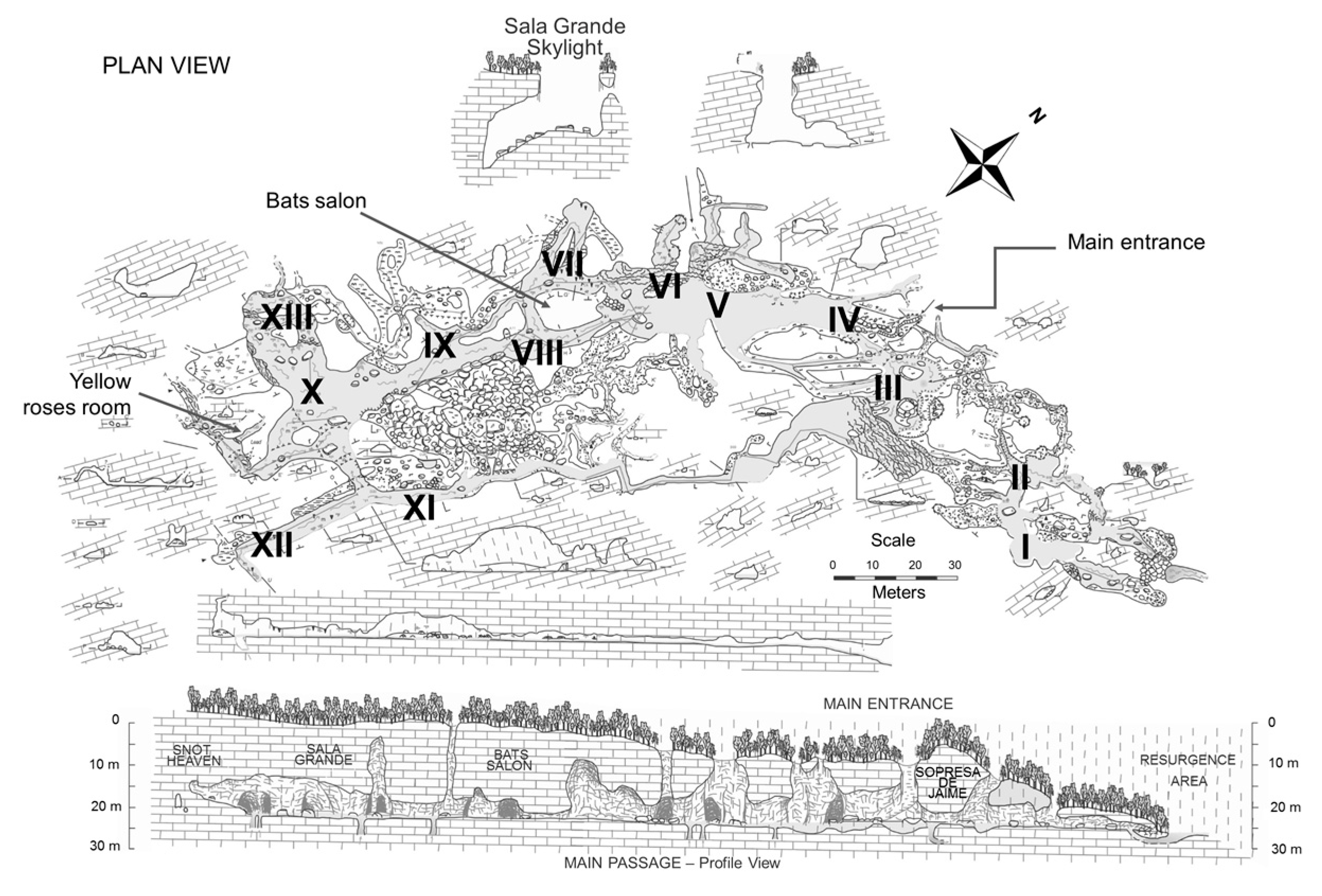
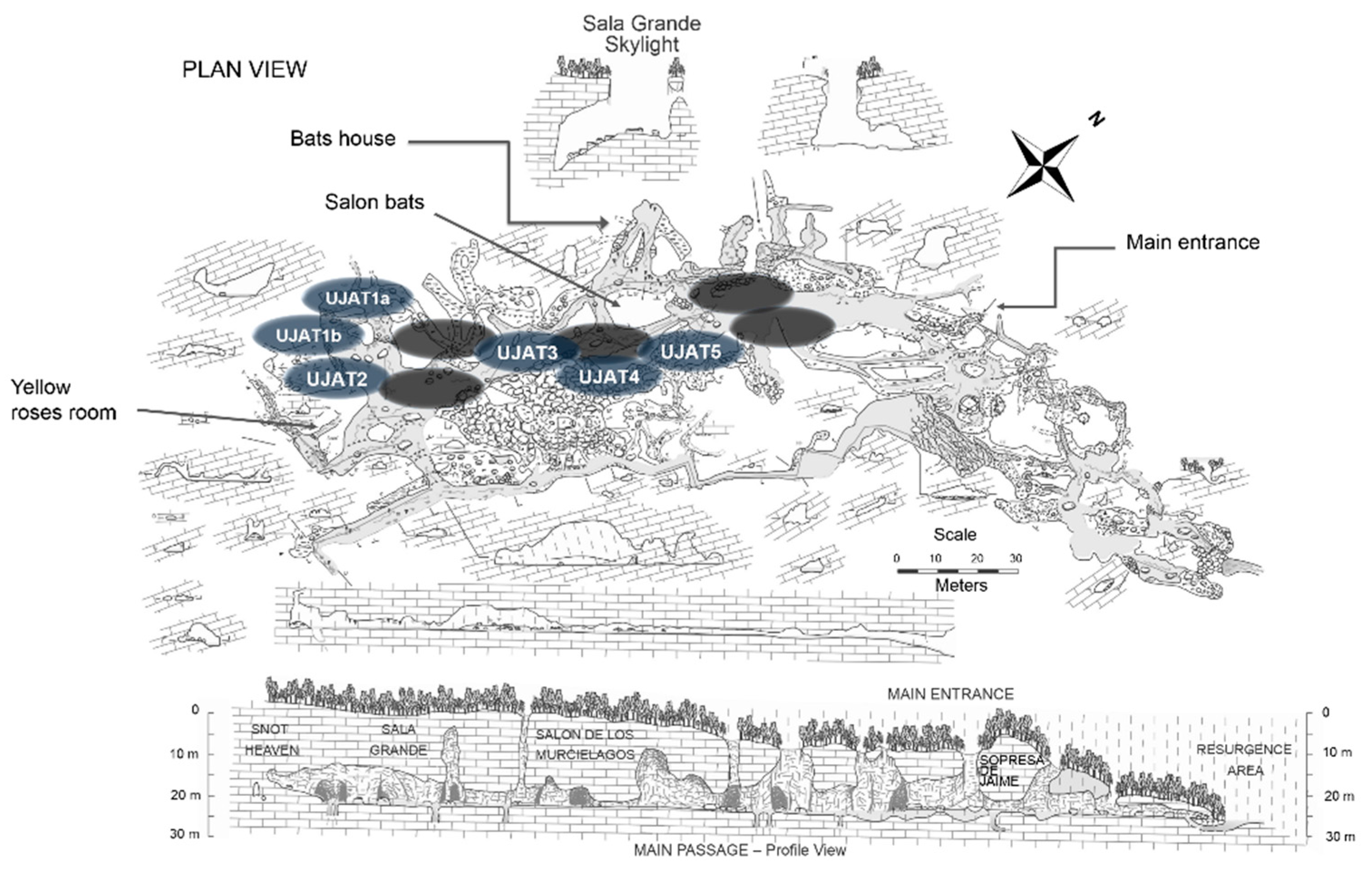
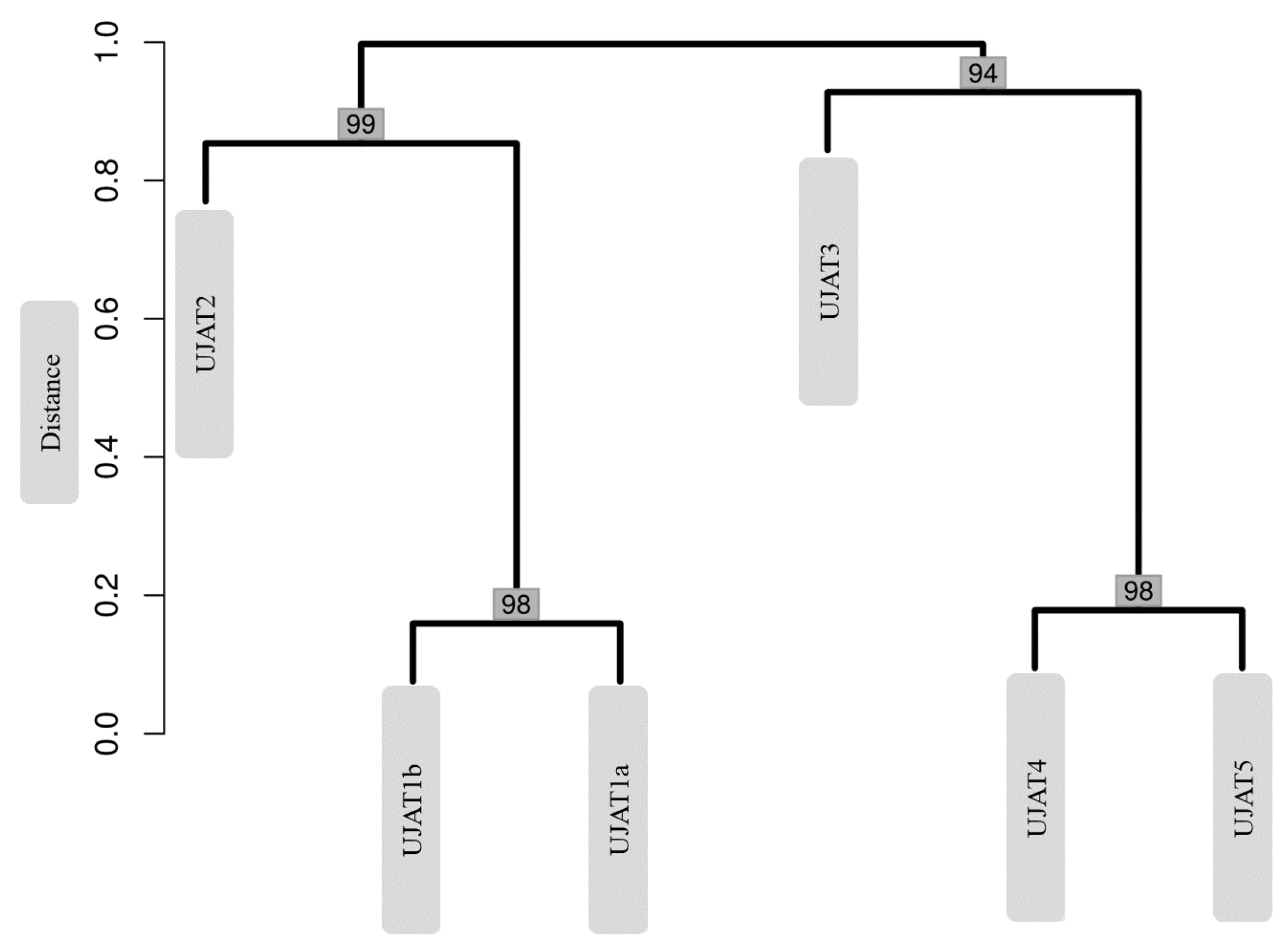
| Sample Type | Selected Sampling Site in Vila Luz cave | Physicochemical Measurements | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | ppmv in the Atmosphere | Concentration (mg L−1) | ||||||||
| CO2 | CH4 | SO42− | Ca2+ | Mg2+ | Na+ | HCO3− | H2S | ||||
| Sediment-water | UJAT1a | 7.0 | 30 | 527 | 2.09 | >960 | >393 | >85.8 | >479 | >1330 | >500 |
| UJAT1b | 28.7 | 960 | ~393 | 85.8 | 479 | 1330 | 500 | ||||
| UJAT2 | 6.8 | 28.6 | 847 | 2.63–2.70 | 75.7–83.8 | 412–465 | |||||
| UJAT3 | 6.9 | 27 | ~396 | 1.87 | <960 | ~387 | |||||
| Biovermiculites | UJAT4 | 7.0 | ~467 | 1.97 | 1310 | 300 | |||||
| Snottites | UJAT5 | 2.5 | 85.8 | 479 | 400 | ||||||
| Reference | This work | [20] | [27] | [37], this work | |||||||
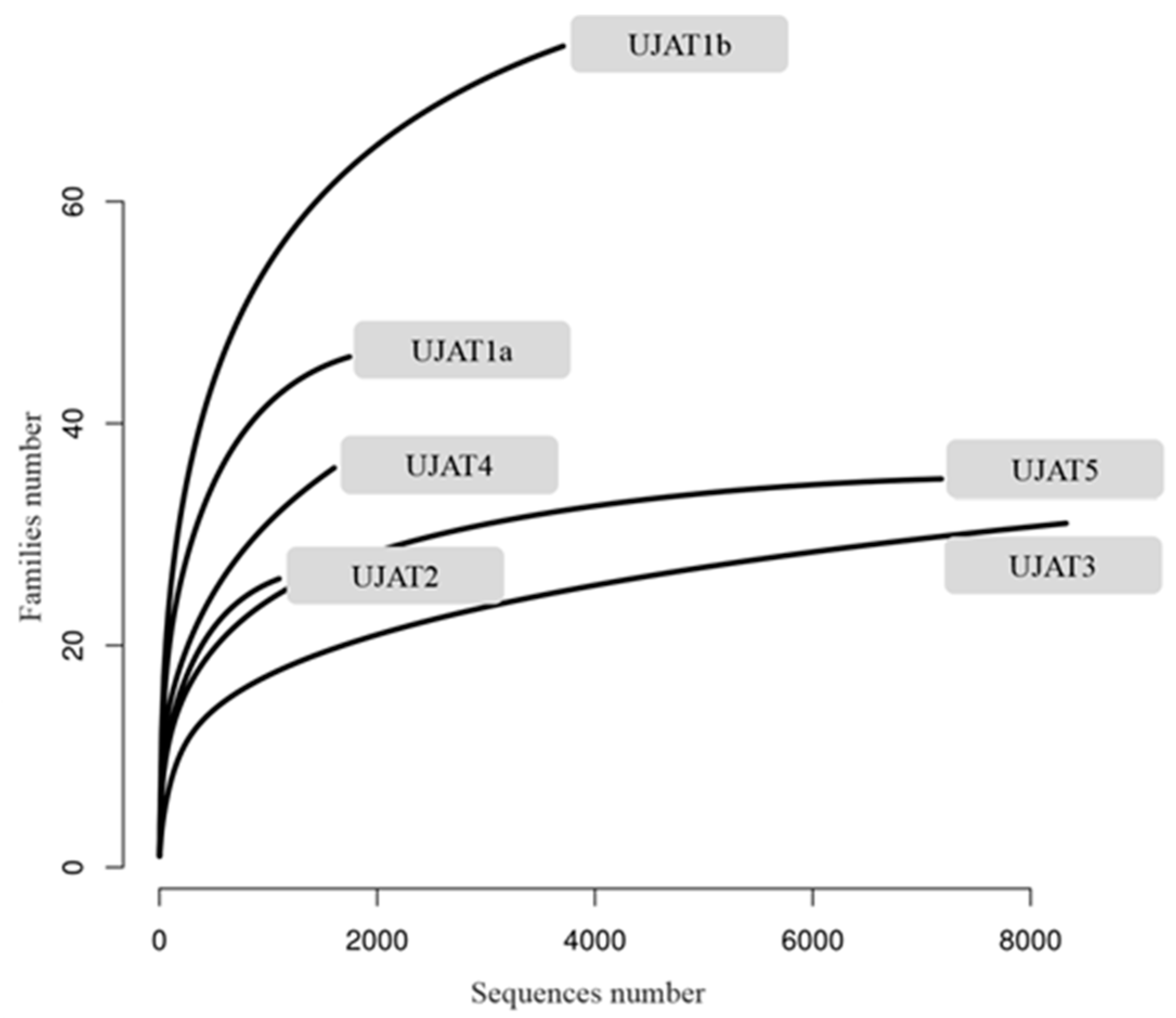
| Sample | Reads | Length (SD) | N Families | Shannon | Chao1 (SE) |
|---|---|---|---|---|---|
| UJAT1a | 1686 | 471.58 (54.77) | 47 | 2.67 | 47.00 (1.77) |
| UJAT1b | 3286 | 428.85 (56.72) | 81 | 3.02 | 81.09 (6.13) |
| UJAT2 | 995 | 424.71 (53.74) | 27 | 2.00 | 27.00 (2.16) |
| UJAT3 | 6842 | 380.98 (46.38) | 36 | 0.73 | 36.06 (7.48) |
| UJAT4 | 1401 | 479.87 (62.95) | 47 | 2.41 | 47.00 (12.47) |
| UJAT5 | 6691 | 419.73 (57.74) | 35 | 1.88 | 35.12 (0.68) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Auria, G.; Artacho, A.; Rojas, R.A.; Bautista, J.S.; Méndez, R.; Gamboa, M.T.; Gamboa, J.R.; Gómez-Cruz, R. Metagenomics of Bacterial Diversity in Villa Luz Caves with Sulfur Water Springs. Genes 2018, 9, 55. https://doi.org/10.3390/genes9010055
D’Auria G, Artacho A, Rojas RA, Bautista JS, Méndez R, Gamboa MT, Gamboa JR, Gómez-Cruz R. Metagenomics of Bacterial Diversity in Villa Luz Caves with Sulfur Water Springs. Genes. 2018; 9(1):55. https://doi.org/10.3390/genes9010055
Chicago/Turabian StyleD’Auria, Giuseppe, Alejandro Artacho, Rafael A. Rojas, José S. Bautista, Roberto Méndez, María T. Gamboa, Jesús R. Gamboa, and Rodolfo Gómez-Cruz. 2018. "Metagenomics of Bacterial Diversity in Villa Luz Caves with Sulfur Water Springs" Genes 9, no. 1: 55. https://doi.org/10.3390/genes9010055
APA StyleD’Auria, G., Artacho, A., Rojas, R. A., Bautista, J. S., Méndez, R., Gamboa, M. T., Gamboa, J. R., & Gómez-Cruz, R. (2018). Metagenomics of Bacterial Diversity in Villa Luz Caves with Sulfur Water Springs. Genes, 9(1), 55. https://doi.org/10.3390/genes9010055





