c-di-AMP: An Essential Molecule in the Signaling Pathways that Regulate the Viability and Virulence of Gram-Positive Bacteria
Abstract
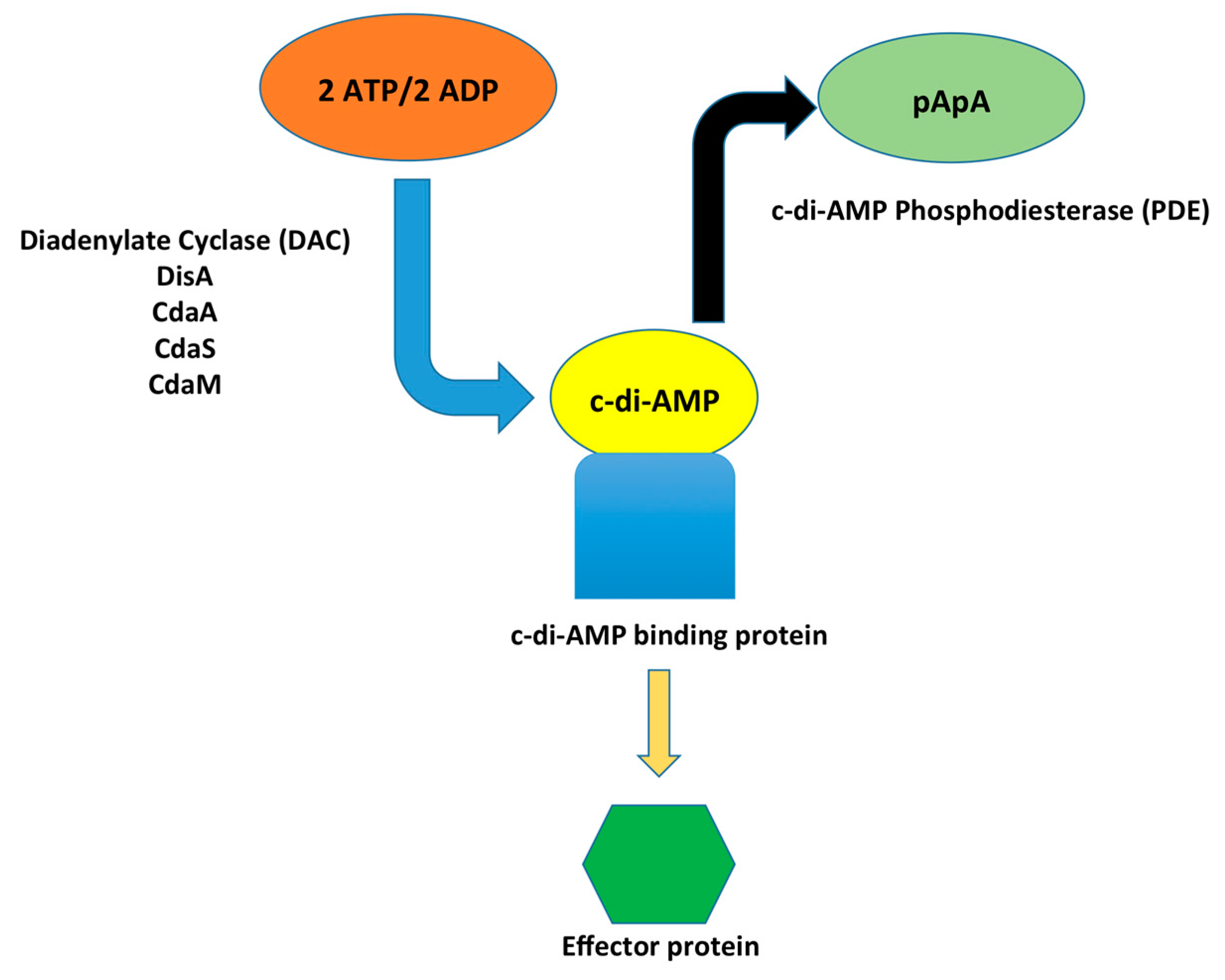
1. Introduction
2. Synthesis of c-di-AMP
2.1. DisA
2.2. CdaA
2.3. CdaS
2.4. CdaM
3. c-di-AMP Degradation
3.1. GdpP
3.2. PgpH
3.3. Pde2
4. c-di-AMP Binding Molecules
4.1. TetR-Family Transcription Factor, DarR
4.2. RCK_C Domain Protein KtrA
4.3. RCK_C Domain Protein CpaA
4.4. Histidine Kinase Protein, KdpD
4.5. PII-Like Signal Transduction Protein, PstA
4.6. KtrA Homolog Protein, CabP
4.7. CabPA and CabPB
4.8. LmPC
4.9. c-di-AMP Binding Riboswitches
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hengge, R.; Grundling, A.; Jenal, U.; Ryan, R.; Yildiz, F. Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J. Bacteriol. 2016, 198, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.L.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013, 42, 305–341. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, C.; Hengge, R. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 2009, 12, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.N.; Choi, P.H.; Sureka, K.; Ledvina, H.E.; Campillo, J.; Tong, L.; Woodward, J.J. Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC. Mol. Microbiol. 2016, 102, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, E.W.; Rall, T.W. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J. Biol. Chem. 1958, 232, 1077–1091. [Google Scholar] [PubMed]
- Rall, T.W.; Sutherland, E.W. Formation of a cyclic adenine ribonucleotide by tissue particles. J. Biol. Chem. 1958, 232, 1065–1076. [Google Scholar] [PubMed]
- McDonough, K.A.; Rodriguez, A. The myriad roles of cyclic AMP in microbial pathogens: From signal to sword. Nat. Rev. Microbiol. 2011, 10, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Gomelsky, M.; Galperin, M.Y. Bacterial second messengers, cGMP and c-di-GMP, in a quest for regulatory dominance. Embo J. 2013, 32, 2421–2423. [Google Scholar] [CrossRef] [PubMed]
- Gomelsky, M. cAMP, c-di-GMP, c-di-AMP and now cGMP: Bacteria use them all! Mol. Microbiol. 2011, 79, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Andrade, W.A.; Firon, A.; Schmidt, T.; Hornung, V.; Fitzgerald, K.A.; Kurt-Jones, E.A.; Trieu-Cuot, P.; Golenbock, D.T.; Kaminski, P.A. Group B Streptococcus degrades cyclic-di-AMP to modulate STING-dependent type I interferon production. Cell HostMicrobe 2016, 20, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Barker, J.R.; Koestler, B.J.; Carpenter, V.K.; Burdette, D.L.; Waters, C.M.; Vance, R.E.; Valdivia, R.H. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. Mbio 2013, 4, e00018-13. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Abbott, J.C.; Burhenne, H.; Kaever, V.; Grundling, A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011, 7, e1002217. [Google Scholar] [CrossRef] [PubMed]
- Gandara, C.; Alonso, J.C. DisA and c-di-AMP act at the intersection between DNA-damage response and stress homeostasis in exponentially growing Bacillus subtilis cells. DNA Repair 2015, 27, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kamegaya, T.; Kuroda, K.; Hayakawa, Y. Identification of a Streptococcus pyogenes SF370 gene involved in production of c-di-AMP. Nagoya J. Med. Sci. 2011, 73, 49–57. [Google Scholar] [PubMed]
- Pham, T.H.; Liang, Z.X.; Marcellin, E.; Turner, M.S. Replenishing the cyclic-di-AMP pool: Regulation of diadenylate cyclase activity in bacteria. Curr. Genet. 2016, 62, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Woodward, J.J.; Iavarone, A.T.; Portnoy, D.A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 2010, 328, 1703–1705. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Campeotto, I.; Jeganathan, T.; Roelofs, K.G.; Lee, V.T.; Grundling, A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 9084–9089. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer-Shaanan, Y.; Wexselblatt, E.; Katzhendler, J.; Yavin, E.; Ben-Yehuda, S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. Embo Rep. 2011, 12, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Mehne, F.M.P.; Schroder-Tittmann, K.; Eijlander, R.T.; Herzberg, C.; Hewitt, L.; Kaever, V.; Lewis, R.J.; Kuipers, O.P.; Tittmann, K.; Stulke, J. Control of the diadenylate cyclase CdaS in Bacillus subtilis: An autoinhibitory domain limits cyclic di-AMP production. J.Biol. Chem. 2014, 289, 21098–21107. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.; Rath, H.; Herzberg, C.; Mader, U.; Stulke, J. Second messenger signaling in Bacillus subtilis: Accumulation of cyclic di-AMP inhibits biofilm formation. Front. Microbiol. 2016, 7, 804. [Google Scholar] [CrossRef] [PubMed]
- Witte, C.E.; Whiteley, A.T.; Burke, T.P.; Sauer, J.D.; Portnoy, D.A.; Woodward, J.J. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. Mbio 2013, 4, e00282-13. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, K.; Sabareesh, V.; Singh, N.; Saigal, K.; Mechold, U.; Sinha, K.M. Two-step synthesis and hydrolysis of cyclic di-AMP in Mycobacterium tuberculosis. PLoS ONE 2014, 9, e86096. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhang, Y.; Bai, G.C.; Zhou, X.D.; Wu, H. Cyclic di-AMP mediates biofilm formation. Mol. Microbiol. 2016, 99, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.Q.; Zheng, X.; Zhou, X.D.; Zeng, J.M.; Ren, Z.; Xu, X.; Cheng, L.; Li, M.Y.; Li, J.Y.; Li, Y.Q. Regulation of oxidative response and extracellular polysaccharide synthesis by a diadenylate cyclase in Streptococcus mutans. Environ. Microbiol. 2016, 18, 904–922. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.L.; Yang, J.; Zarrella, T.M.; Zhang, Y.; Metzger, D.W.; Bai, G.C. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J. Bacteriol. 2014, 196, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Kang, S.O. Streptococcus pyogenes c-di-AMP phosphodiesterase, GdpP, influences SpeB processing and virulence. PLoS ONE 2013, 8, e69425. [Google Scholar] [CrossRef] [PubMed]
- Bin, D.; Ji, W.H.; An, H.T.; Shi, Y.B.; Huang, Q.Q.; Cheng, Y.Q.; Fu, Q.; Wang, H.G.; Yan, Y.X.; Sun, J.H. Functional analysis of c-di-AMP phosphodiesterase, GdpP, in Streptococcus suis serotype 2. Microbiol. Res. 2014, 169, 749–758. [Google Scholar]
- Blötz, C.; Treffon, K.; Kaever, V.; Schwede, F.; Hammer, E.; Stülke, J. Identification of the components involved in cyclic di-AMP signaling in Mycoplasma pneumoniae. Front. Microbiol. 2017, 8, 1328. [Google Scholar] [CrossRef] [PubMed]
- Witte, G.; Hartung, S.; Buttner, K.; Hopfner, K.P. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 2008, 30, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Temeng, C.; Zhou, J.; Zheng, Y.; Su, J.; Sintim, H.O. Cyclic dinucleotide (c-di-GMP, c-di-AMP, and cGAMP) signalings have come of age to be inhibited by small molecules. Chem. Commun. 2016, 52, 9327–9342. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, A.T.; Pollock, A.J.; Portnoy, D.A. The PAMP c-di-AMP Is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe 2015, 18, 132. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Grundling, A. Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 2013, 11, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.N.; Woodward, J.J. Too much of a good thing: Regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr. Opin. Microbiol. 2016, 30, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Commichau, F.M.; Dickmanns, A.; Gundlach, J.; Ficner, R.; Stulke, J. A jack of all trades: The multiple roles of the unique essential second messenger cyclic di-AMP. Mol. Microbiol. 2015, 97, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Helmann, J. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol. 2012, 83, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yang, J.; Zhou, X.; Ding, X.; Eisele, L.E.; Bai, G. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS ONE 2012, 7, e35206. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.; Mehne, F.M.P.; Herzberg, C.; Kampf, J.; Valerius, O.; Kaever, V.; Stulke, J. An essential poison: Synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J. Bacteriol. 2015, 197, 3265–3274. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.; Dickmanns, A.; Neumann, P.; Gunka, K.; Arens, J.; Kaever, V.; Stulke, J.; Ficner, R.; Commichau, F.M. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J. Biol. Chem. 2015, 290, 6596–6606. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Bowman, L.; Willis, A.R.; Kaever, V.; Grundling, A. Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J. Biol. Chem. 2015, 290, 5826–5839. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Pham, T.H.; Nhiep, T.H.; Vu, N.M.; Marcellin, E.; Chakrabortti, A.; Wang, Y.; Waanders, J.; Lo, R.; Huston, W.M.; et al. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol. Microbiol. 2016, 99, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.L.; Yang, J.; Eisele, L.E.; Underwood, A.J.; Koestler, B.J.; Waters, C.M.; Metzger, D.W.; Bai, G.C. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J. Bacteriol. 2013, 195, 5123–5132. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.N.; Luo, S.K.; Pensinger, D.; Sauer, J.D.; Tong, L.; Woodward, J.J. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. USA 2015, 112, E747–E756. [Google Scholar] [CrossRef] [PubMed]
- Rao, F.; See, R.Y.; Zhang, D.W.; Toh, D.C.; Ji, Q.; Liang, Z.X. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 2010, 286, 29441. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Helmann, J. A σD-dependent antisense transcript modulates expression of the cyclic-di-AMP hydrolase GdpP in Bacillus subtilis. Microbiology 2012, 158, 2732–2741. [Google Scholar]
- Tan, E.; Rao, F.; Pasunooti, S.; Pham, T.H.; Soehano, I.; Turner, M.S.; Liew, C.W.; Lescar, J.; Pervushin, K.; Liang, Z.X. Solution structure of the PAS domain of a thermophilic YybT protein homolog reveals a potential ligand-binding site. J. Biol. Chem. 2013, 288, 11949–11959. [Google Scholar] [CrossRef] [PubMed]
- Sureka, K.; Choi, P.H.; Precit, M.; Delince, M.; Pensinger, D.A.; Huynh, T.N.; Jurado, A.R.; Goo, Y.A.; Sadilek, M.; Iavarone, A.T.; et al. The cyclic dinucleotide c-di-AMP Is an allosteric regulator of metabolic enzyme function. Cell 2014, 158, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Cron, L.E.; Stol, K.; Burghout, P.; van Seim, S.; Simonetti, E.R.; Bootsma, H.J.; Hermans, P.W.M. Two DHH subfamily 1 proteins contribute to pneumococcal virulence and confer protection against pneumococcal disease. Infect. Immun. 2012, 80, 2974. [Google Scholar]
- Bowman, L.; Zeden, M.S.; Schuster, C.F.; Kaever, V.; Grundling, A. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J. Biol. Chem. 2016, 291, 26970–26986. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Wang, F.; Liu, S.; Zhu, D.; Cong, H.; Gao, F.; Li, B.; Wang, H.; Lin, Z.; Liao, J.; et al. Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase. J. Biol. Chem. 2016, 291, 14386–14387. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Luo, Y.; Zheng, C.; Yin, K.; Ali, M.K.; Li, X.; He, J. Functional analysis of a c-di-AMP-specific phosphodiesterase MsPDE from Mycobacterium smegmatis. Int. J. Biol. Sci. 2015, 11, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Bai, Y.L.; Zhang, Y.; Gabrielle, V.D.; Jin, L.; Bai, G.C. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol. Microbiol. 2014, 93, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Zhang, J.J.; Fang, X.; Lawlis, G.B.; Troxell, B.; Zhou, Y.; Gomelsky, M.; Lou, Y.; Yang, X.F. DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect. Immun. 2014, 82, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, W.H.; He, Z.G. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J. Biol. Chem. 2013, 288, 3085–3096. [Google Scholar] [CrossRef] [PubMed]
- Sudarsan, N.; Lee, E.R.; Weinberg, Z.; Moy, R.H.; Kim, J.N.; Link, K.H.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 2008, 321, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.M.; Patel, D.J. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat. Chem. Biol. 2014, 10, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Youn, S.J.; Kim, S.O.; Ko, J.; Lee, J.O.; Choi, B.S. Structural studies of potassium transport protein KtrA regulator of conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c-di-AMP). J. Biol. Chem. 2015, 290, 16393–16402. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Serganov, A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat. Chem. Biol. 2014, 10, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.P.; Ferre-D’Amare, A.R. Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA. Embo J. 2014, 33, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.M.; Pham, T.H.; Lei, L.; Dou, J.C.; Soomro, A.H.; Beatson, S.A.; Dykes, G.A.; Turner, M.S. Heat resistance and salt hypersensitivity in Lactococcus lactis due to spontaneous mutation of llmg_1816 (gdpP) induced by high-temperature growth. Appl. Environ. Microbiol. 2012, 78, 7753–7759. [Google Scholar] [CrossRef] [PubMed]
- Freeman, Z.N.; Dorus, S.; Waterfield, N.R. The KdpD/KdpE two-component system: Integrating K+ homeostasis and virulence. PLoS Pathog. 2013, 9, e1003201. [Google Scholar] [CrossRef] [PubMed]
- Ballal, A.; Basu, B.; Apte, S.K. The Kdp-ATPase system and its regulation. J. Biosci. 2007, 32, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.H.; Sureka, K.; Woodward, J.J.; Tong, L. Molecular basis for the recognition of cyclic-di-AMP by PstA, a PII-like signal transduction protein. Microbiologyopen 2015, 4, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Pires, R.S.; Szollosi, A.; Morais-Cabral, J.H. The structure of the KtrAB potassium transporter. Nature 2013, 496, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.H.; Liang, J.M.; Yang, J.G.; Shih, M.S.; Tu, Z.L.; Wang, Y.C.; Sun, X.H.; Hu, N.J.; Liang, Z.X.; Dow, J.M.; et al. Structural insights into the distinct binding mode of cyclic di-AMP with SaCpaA_RCK. Biochemistry 2015, 54, 4936–4951. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, J.A.; Schramke, H.; Zhang, Y.; Tosi, T.; Dehbi, A.; Jung, K.; Grundling, A. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter. J. Bacteriol. 2015, 198, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Campeotto, I.; Zhang, Y.; Mladenov, M.G.; Freemont, P.S.; Grundling, A. Complex structure and biochemical characterization of the Staphylococcus aureus cyclic diadenylate monophosphate (c-di-AMP)-binding protein PstA, the founding member of a new signal transduction protein family. J. Biol. Chem. 2015, 290, 2888–2901. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.; Dickmanns, A.; Schroder-Tittmann, K.; Neumann, P.; Kaesler, J.; Kampf, J.; Herzberg, C.; Hammer, E.; Schwede, F.; Kaever, V.; et al. Identification, characterization, and structure analysis of the cyclic di-AMP-binding PII-like signal transduction protein DarA. J. Biol. Chem. 2015, 290, 3069–3080. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Corbino, K.A.; Winkler, W.C.; Nahvi, A.; Mandal, M.; Collins, J.; Lee, M.; Roth, A.; Sudarsan, N.; Jona, I.; et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA 2004, 101, 6421–6426. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.Y.; Fedor, M.J. The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat. Chem. Biol. 2012, 8, 963–965. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Sudarsan, N.; Furukawa, K.; Weinberg, Z.; Wang, J.X.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013, 9, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.; Herzberg, C.; Kaever, V.; Gunka, K.; Hoffmann, T.; Weiß, M.; Gibhardt, J.; Thurmer, A.; Hertel, D.; Daniel, R.; et al. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci. Signal. 2017, 10, eaal3011. [Google Scholar] [CrossRef] [PubMed]
| Bacterium | Function of c-di-AMP | c-di-AMP Synthesis Enzyme | c-di-AMP Degrading Enzyme | Phenotype Involved in an Altered Level of c-di-AMP | Ref. |
|---|---|---|---|---|---|
| Bacillus subtilis | DisA binds to DNA and maintains DNA integrity. CdaS regulates sporulation. CdaA regulates cell wall synthesis and ion channel homeostasis. | DisA, CdaA, and CdaS | GdpP and PgpH | DisA mutation: decreased DNA integrity CdaA mutation: impaired potassium ion channel system, weakened cell wall, increased resistance to antibiotics CdaS mutation: delayed sporulation. | [17,18,19,20] |
| Listeria monocytogenes | Regulates cell wall homeostasis, resistance to acid, and carbon metabolism. | CdaA (DacA) | PdeA (GdpP homolog) and PgpH | Phosphodiesterase (PDE mutation: cell wall defects, increased resistance to antibiotics, low survival rate, sensitivity towards acid stress, altered interferon-ß stimulation in host cells. | [21] |
| Mycobacterium tuberculosis | Functions are not fully understood yet, but DisA is predicted to be involved in DNA repair. | MtbDisA (DisA ortholog) | MtbPDE (Pde2 ortholog) | PDE mutation: reduced virulence. | [22] |
| Staphylococcus aureus | Regulates cell wall synthesis, cell size, and potassium ion channel homeostasis. | CdaA | GdpP and Pde2 ortholog | gdpP deletion: smaller cell size, increased peptidoglycan cross-linking, increased resistance against cell wall and membrane targeting antibiotics, impaired potassium ion channel system. | [17] |
| Streptococcus mutans | Regulates biofilm formation by binding to receptor proteins. | CdaA | PdeA (GdpP ortholog) and Pde2 | cda deletion: Increased sensitivity to hydrogen peroxide and enhanced polysaccharide synthesis. pdeA deletion: Increased biofilm formation. | [23,24] |
| Streptococcus pneumoniae | Maintains potassium ion channel homeostasis. | CdaA | GdpP and Pde2 | PDE mutation: Impaired ability of long chain formation, decreased growth, and imbalance in the potassium ion channel. | [25] |
| Streptococcus pyogenes | Regulates cell wall homeostasis and virulence gene expression. | CdaA (SpyDacA) | GdpP and Pde2 ortholog | gdpP deletion: Impaired biogenesis of SpeB, decreased virulence and increased antibiotic resistance. | [26] |
| Streptococcus suis (SS2) | Promotes biofilm formation and increases virulence. | CdaA | GdpP and Pde2 ortholog | gdpP deletion: Reduced growth and reduced biofilm formation. | [27] |
| Mycoplasma pneumoniae | Predicted to regulate potassium import through binding of KtrC. | CdaM | PdeM | cdaM and pdeM: essential for growth. | [28] |
| c-di-AMP Receptor Proteins (Species Originally Identified in) | Location in the Cell | Protein Structure and Functional Domains | Protein Function | Phenotypes by Deletion or Overexpression of the Genes | References |
|---|---|---|---|---|---|
| DarR (Mycobacterium smegmatis,) | Cytoplasmic protein | DarR contains two domains: a C-terminal QacR-like domain, and an N-terminal TetR-like helix-turn-helix domain. The binding site of c-di-AMP has not been identified yet. | Transcriptional repressor for genes involved in ion transport, membrane lipid homeostasis, and stress response. | Deletion causes larger cell size. Overexpression is toxic to cells and causes reduced fatty acid metabolism. | [38,55] |
| KtrA (S. aureus) | Cytoplasmic protein bound to the integral membrane protein KtrB | KtrA possesses two RCK domains, RCK_N and RCK_C. c-di-AMP binds to RCK_C. | KtrA-KtrB complex regulates the potassium ion channel opening and closing by changing their conformation following c-di-AMP binding. | Deletion causes sensitivity to osmotic stress and requires high levels of potassium for growth. | [17,32,58,61] |
| CpaA (S. aureus) | Integral membrane protein | Twelve membrane-spanning region, RCK_N and RCK_C domain c-di-AMP binds to RCK_C. | A putative proton antiporter in the cell where intracellular protons are exchanged with potassium or sodium ions. | Not studied yet. | [17,32] |
| KdpD (S. aureus) | Integral membrane protein | KdpD is a histidine kinase in a TCS. | Regulates a P-type ATP- dependent high-affinity potassium uptake system. | Deletion leads to low virulence and less survival. | [32,35,62,63] |
| PstA (S. aureus) | Cytoplasmic protein | PII-like signal transduction protein with an unknown domain DUF970. | Unknown. | Not studied yet. | [64] |
| CabP (S. pneumonaie) | Cytoplasmic protein | An octameric protein belonging to the Trk family Homolog of KtrA. Binds to the ortholog of KtrB SPD_0076. | A member of the potassium ion transporter. | CabP mutant exhibits low potassium ion uptake. | [25] |
| CabPA (S. mutans) | Cytoplasmic protein | Trk family.protein. | Binds to VicR, facilitates biofilm formation. | Reduces biofilm formation ability. | [23] |
| CabPB (S. mutans) | Cytoplasmic protein | Trk family protein. | Unknown. | CabPB mutant strains have not been studied yet. | [23] |
| LmPC (L. monocytogenes) | Cytoplasmic protein | Pyruvate carboxylase family, c-di-AMP binds to the dimer interface. | Pyruvate carboxylase: ATP-dependent carboxylation of pyruvate to oxaloacetate. | Causes metabolic imbalance, lysis of bacterial cells during infection. | [48] |
| CbpA, CbpB (L. monocytogenes) | Soluble proteins | Function unknown. | Unknown. | Not studied yet. | [48] |
| NrdR (L. monocytogenes) | Cytoplasmic protein | Transcriptional repressor. | Transcriptional regulator. | Not studied yet. | [48] |
| ydaO Riboswitches (B. subtilis) | Cytoplasmic RNA molecules | The regulatory RNA molecules contain a ligand-sensing domain and an expression platform. c-di-AMP binds to the ligand-sensing domain. | Regulates ion channels, responds to osmotic stress, and facilitates cell wall metabolism and sporulation. | Not essential. | [32,35,45,56,57,58,59,60] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fahmi, T.; Port, G.C.; Cho, K.H. c-di-AMP: An Essential Molecule in the Signaling Pathways that Regulate the Viability and Virulence of Gram-Positive Bacteria. Genes 2017, 8, 197. https://doi.org/10.3390/genes8080197
Fahmi T, Port GC, Cho KH. c-di-AMP: An Essential Molecule in the Signaling Pathways that Regulate the Viability and Virulence of Gram-Positive Bacteria. Genes. 2017; 8(8):197. https://doi.org/10.3390/genes8080197
Chicago/Turabian StyleFahmi, Tazin, Gary C. Port, and Kyu Hong Cho. 2017. "c-di-AMP: An Essential Molecule in the Signaling Pathways that Regulate the Viability and Virulence of Gram-Positive Bacteria" Genes 8, no. 8: 197. https://doi.org/10.3390/genes8080197
APA StyleFahmi, T., Port, G. C., & Cho, K. H. (2017). c-di-AMP: An Essential Molecule in the Signaling Pathways that Regulate the Viability and Virulence of Gram-Positive Bacteria. Genes, 8(8), 197. https://doi.org/10.3390/genes8080197






