Requirements for Efficient Thiosulfate Oxidation in Bradyrhizobium diazoefficiens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
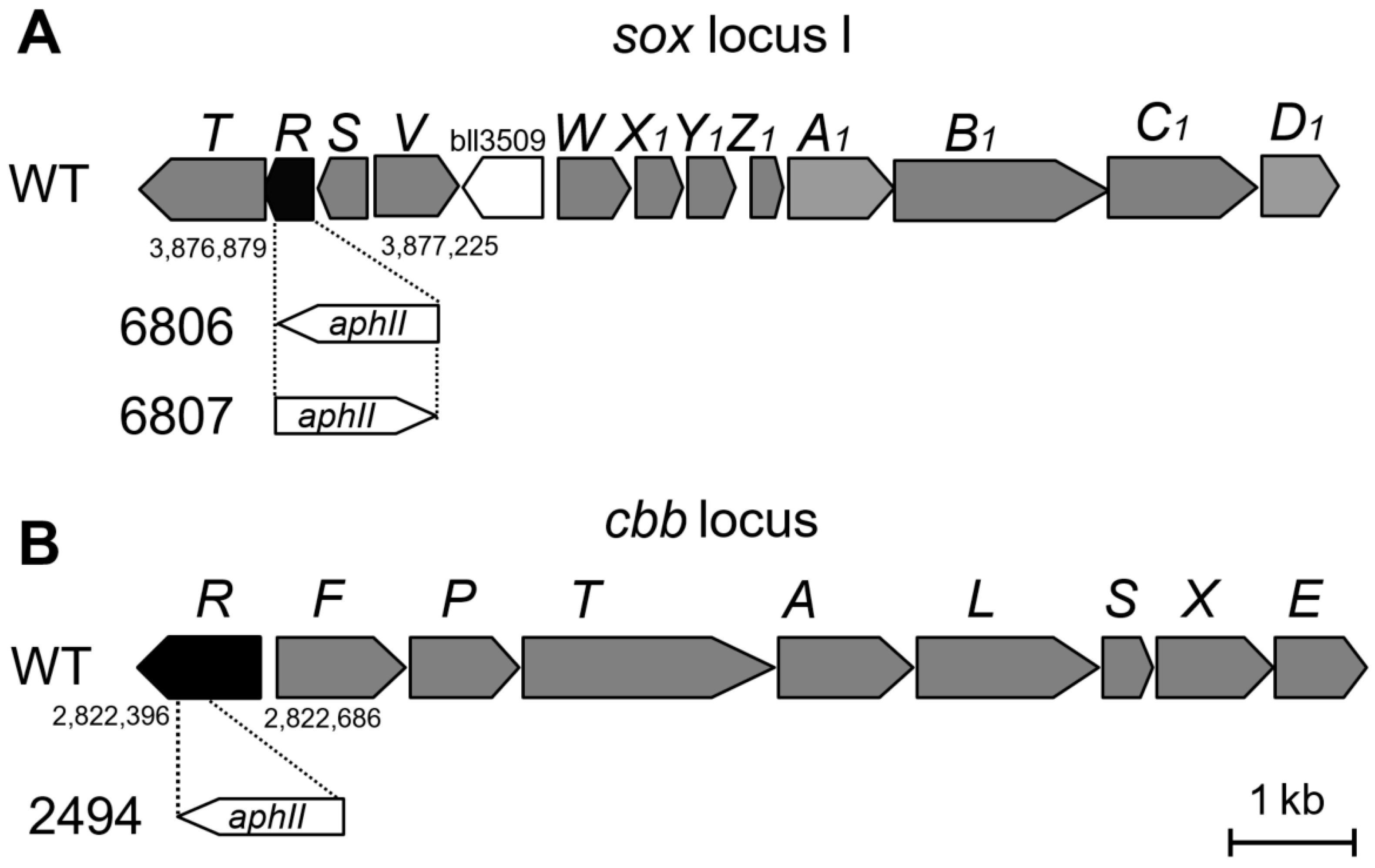
2.2. Construction of soxR and cbbR Deletion Mutants
2.3. Expression of sox and cbb Genes
2.4. Analysis of Thiosulfate, Succinate and Fumarate
3. Results and Discussion
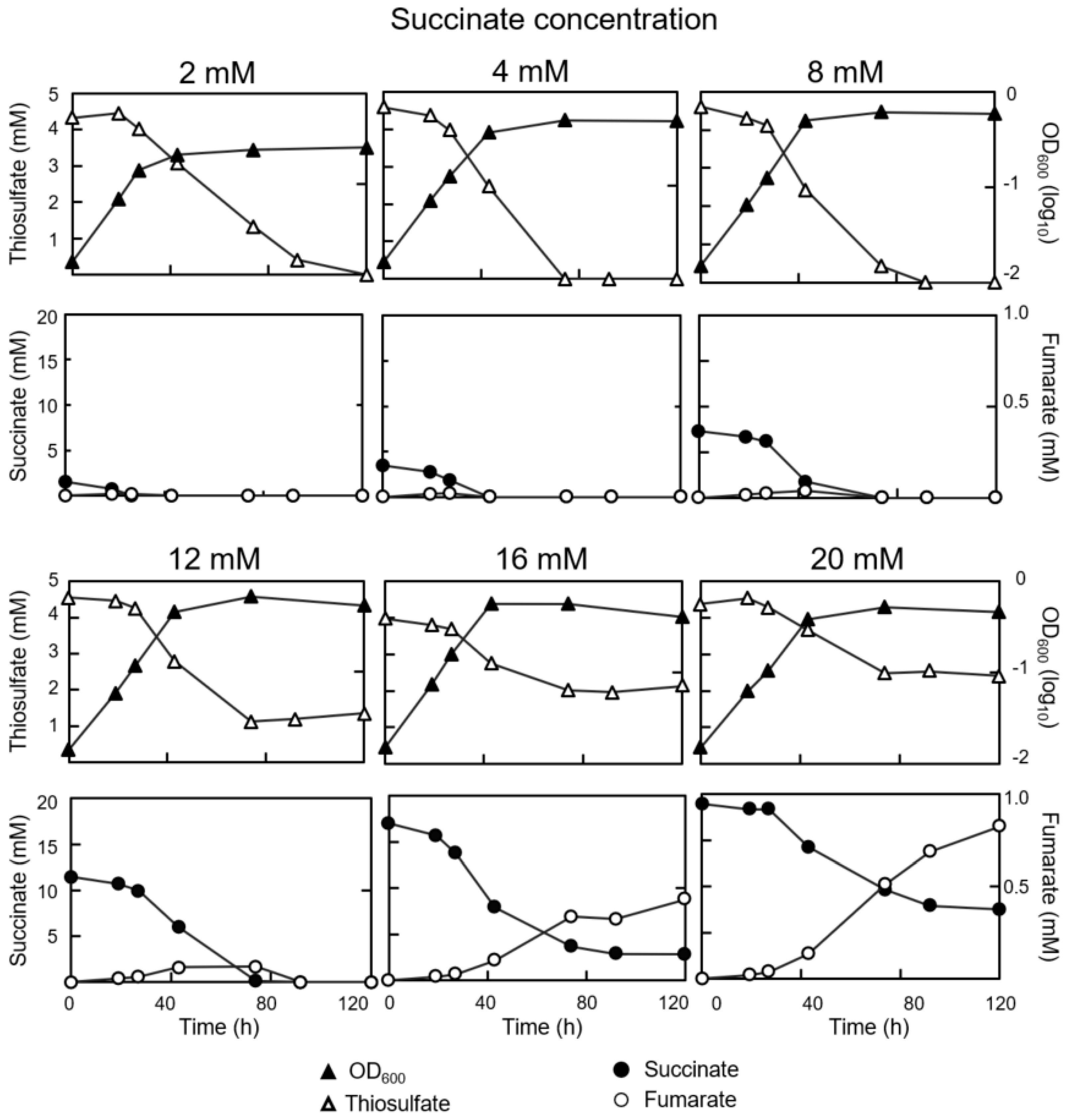
3.1. Concurrent Consumption of Thiosulfate and Succinate during Mixotrophic Growth
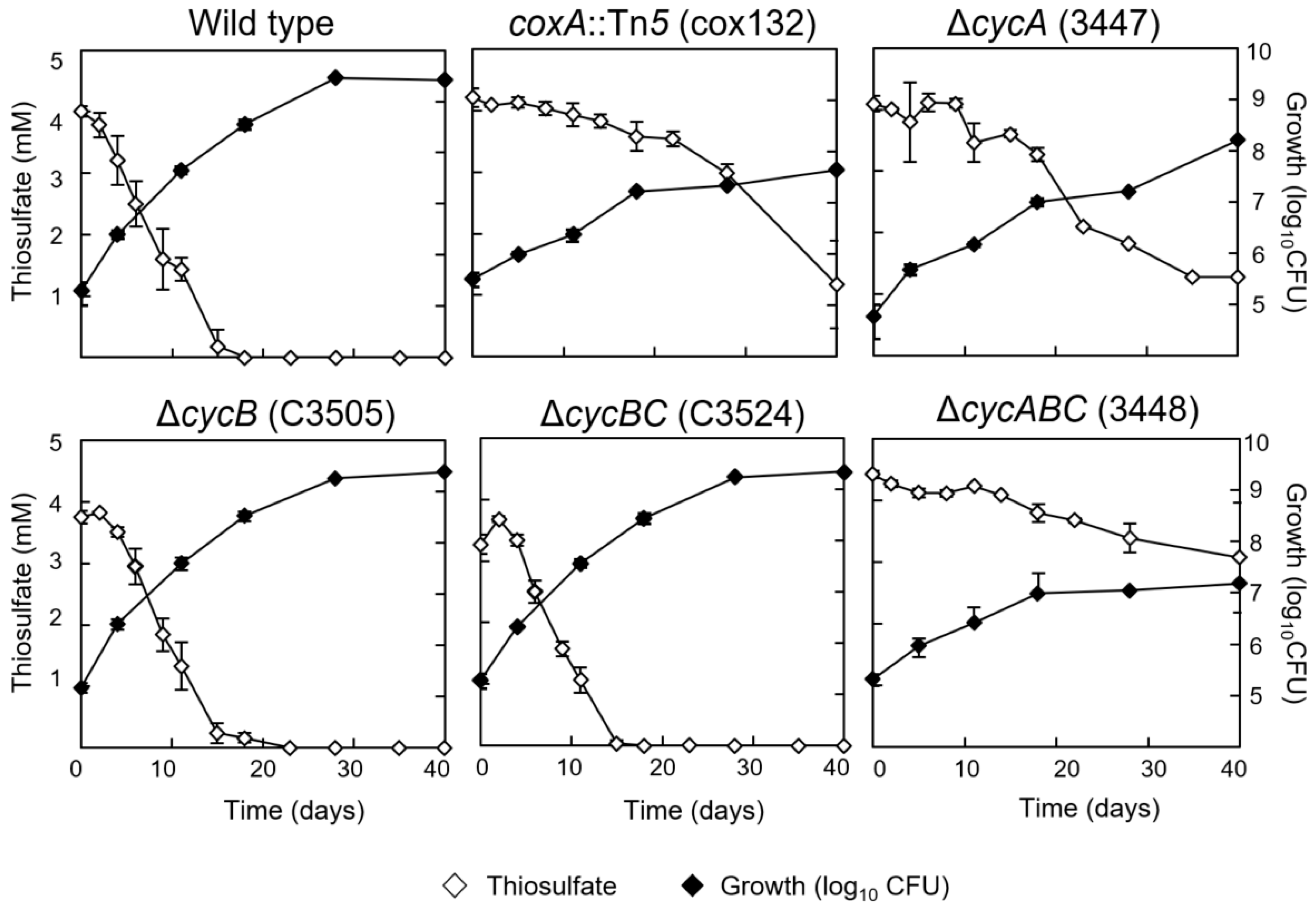
3.2. Chemolithoautotrophic Growth Partly Depends on Cytochrome c550 and the aa3-Type Cytochrome Oxidase
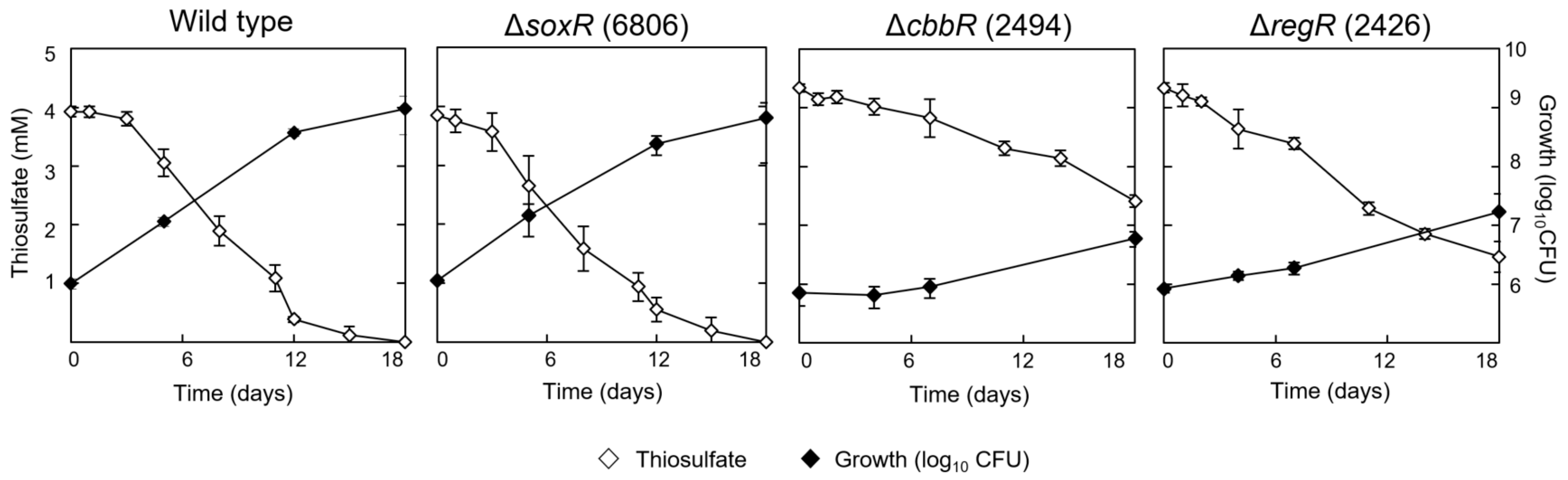
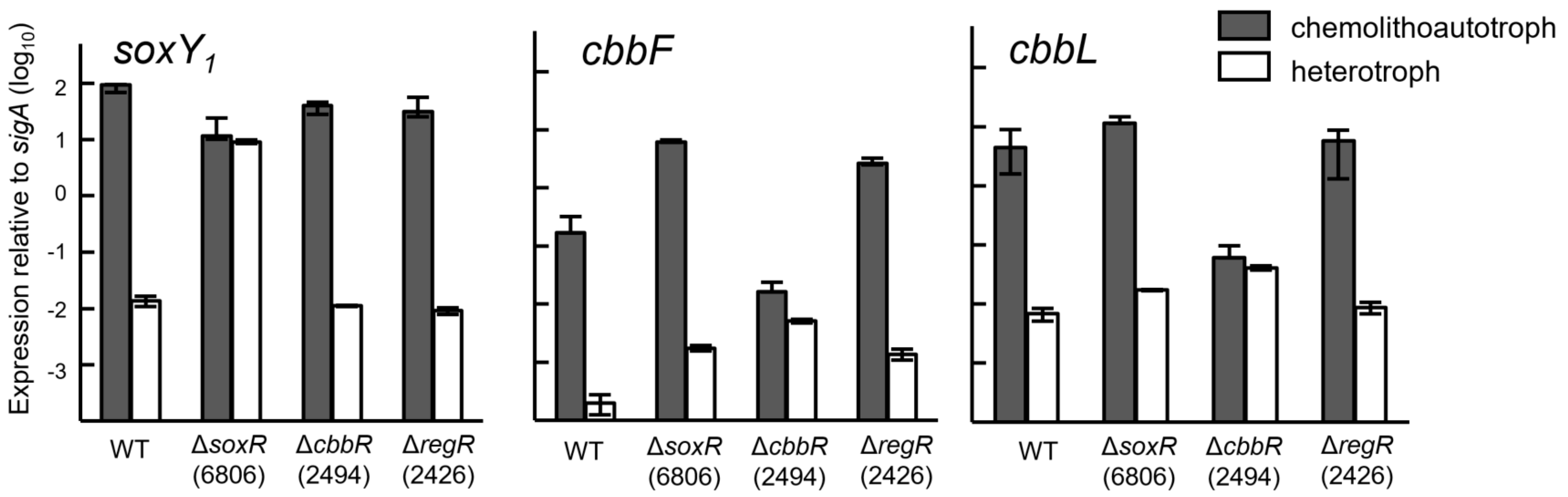
3.3. Involvement of CbbR and RegR in the Regulation of Chemolithotrophic Thiosulfate Oxidation
3.4. Expression of Cbb and Sox Genes during Chemolithoautotrophic Growth
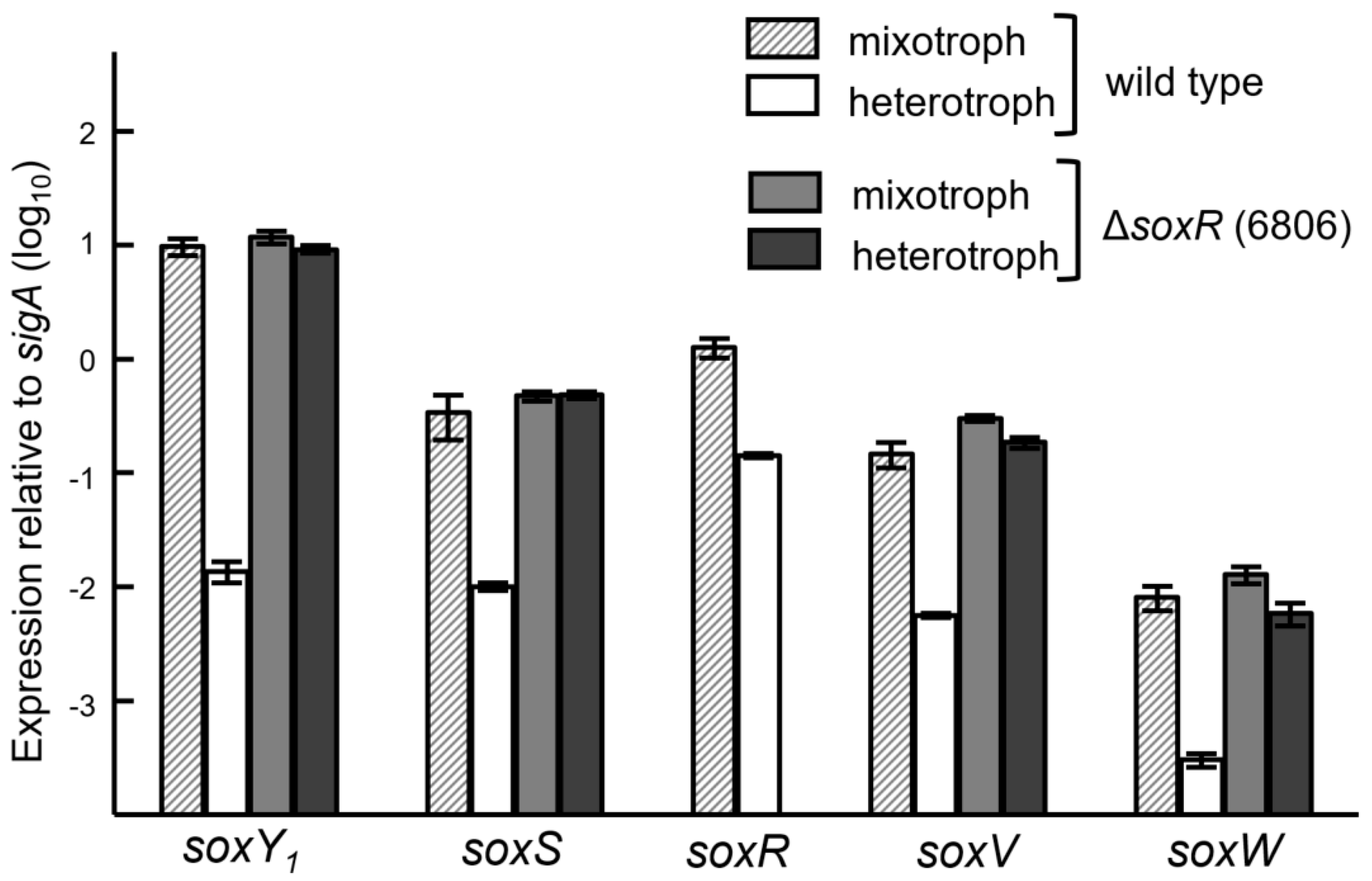
3.5. Expression of the Sox Genes and Evidence for SoxR as a Negative Regulator
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hanus, F.J.; Maier, R.J.; Evans, H.J. Autotrophic growth of H2-uptake-positive strains of Rhizobium japonicum in an atmosphere supplied with hydrogen gas. Proc. Natl. Acad. Sci. USA 1979, 76, 1788–1792. [Google Scholar] [CrossRef] [PubMed]
- Lorite, M.J.; Tachil, J.; Sanjuán, J.; Meyer, O.; Bedmar, E.J. Carbon monoxide dehydrogenase activity in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 2000, 66, 1871–1876. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S.; Eda, S.; Ikeda, S.; Mitsui, H.; Minamisawa, K. Thiosulfate-dependent chemolithoautotrophic growth of Bradyrhizobium japonicum. Appl. Environ. Microbiol. 2010, 76, 2402–2409. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S.; Eda, S.; Sugawara, C.; Mitsui, H.; Minamisawa, K. The cbbL gene is required for thiosulfate-dependent autotrophic growth of Bradyrhizobium japonicum. Microbes Environ. 2010, 25, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Minamisawa, K.; Uchiumi, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Iriguchi, M.; Kawashima, K.; et al. Complete genome sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.G. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 1998, 39, 235–289. [Google Scholar] [PubMed]
- Friedrich, C.G.; Bardischewsky, F.; Rother, D.; Quentmeier, A.; Fischer, J. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 2005, 8, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, W.; Dam, B. Biochemistry and molecular biology of lithoautotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 2009, 33, 999–1043. [Google Scholar] [CrossRef] [PubMed]
- Bühler, D.; Rossmann, R.; Landolt, S.; Balsiger, S.; Fischer, H.M.; Hennecke, H. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. J. Biol. Chem. 2010, 285, 15704–15713. [Google Scholar] [CrossRef] [PubMed]
- Rother, D.; Orawski, G.; Bardischewsky, F.; Friedrich, C.G. SoxRS-mediated regulation of chemotrophic sulfur oxidation in Paracoccus pantotrophus. Microbiology 2005, 151, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Chatterjee, S.; Dam, B.; Roy, P.; Das, G.S.K. The dimeric repressor SoxR binds cooperatively to the promoter(s) regulating expression of the sulfur oxidation (sox) operon of Pseudaminobacter salicylatoxidans KCT001. Microbiology 2007, 153, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.L.; Tabita, F.R. Nucleotide sequence and functional analysis of cbbR, a positive regulator of the Calvin cycle operons of Rhodobacter sphaeroides. J. Bacteriol. 1993, 175, 5778–5784. [Google Scholar] [CrossRef] [PubMed]
- Van den Bergh, E.R.; Dijkhuizen, L.; Meijer, W.G. CbbR, a LysR-type transcriptional activator, is required for expression of the autotrophic CO2 fixation enzymes of Xanthobacter flavus. J. Bacteriol. 1993, 175, 6097–6104. [Google Scholar] [CrossRef] [PubMed]
- Windhövel, U.; Bowien, B. Identification of cfxR, an activator gene of autotrophic CO2 fixation in Alcaligenes eutrophus. Mol. Microbiol. 1991, 5, 2695–2705. [Google Scholar] [CrossRef] [PubMed]
- Bauer, E.; Kaspar, T.; Fischer, H.M.; Hennecke, H. Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J. Bacteriol. 1998, 180, 3853–3863. [Google Scholar] [PubMed]
- Lindemann, A.; Moser, A.; Pessi, G.; Hauser, F.; Friberg, M.; Hennecke, H.; Fischer, H.M. New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J. Bacteriol. 2007, 189, 8928–8943. [Google Scholar] [CrossRef] [PubMed]
- Elsen, S.; Swem, L.R.; Swem, D.L.; Bauer, C.E. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 2004, 68, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Regensburger, B.; Hennecke, H. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 1983, 135, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Delamuta, J.R.; Ribeiro, R.A.; Ormeño-Orrillo, E.; Melo, I.S.; Martinez-Romero, E.; Hungria, M. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3342–3351. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Bott, M.; Bolliger, M.; Hennecke, H. Genetic analysis of the cytochrome c-aa3 branch of the Bradyrhizobium japonicum respiratory chain. Mol. Microbiol. 1990, 4, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- Bott, M.; Thöny-Meyer, L.; Loferer, H.; Rossbach, S.; Tully, R.E.; Keister, D.; Appleby, C.A.; Hennecke, H. Bradyrhizobium japonicum cytochrome c550 is required for nitrate respiration but not for symbiotic nitrogen fixation. J. Bacteriol. 1995, 177, 2214–2217. [Google Scholar] [CrossRef] [PubMed]
- Rossbach, S.; Loferer, H.; Acuña, G.; Appleby, C.A.; Hennecke, H. Cloning, sequencing, and mutational analysis of the cytochrome c552 gene (cycB) from Bradyrhizobium japonicum strain 110. FEMS Microbiol. Lett. 1991, 83, 145–152. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Alexeyev, M.F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques 1995, 18, 52–54. [Google Scholar] [PubMed]
- Fischer, H.M.; Babst, M.; Kaspar, T.; Acuña, G.; Arigoni, F.; Hennecke, H. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 1993, 12, 2901–2912. [Google Scholar] [PubMed]
- Norrander, J.; Kempe, T.; Messing, J. Construction of improved M13 vectors using oligodeoxynucleotide mutagenesis. Gene 1983, 26, 101–106. [Google Scholar] [CrossRef]
- Hahn, M.; Hennecke, H. Localized mutagenesis in Rhizobium japonicum. Mol. Gen. Genet. 1984, 193, 46–52. [Google Scholar] [CrossRef]
- Schneider, K.; Peyraud, R.; Kiefer, P.; Christen, P.; Delmotte, N.; Massou, S.; Portais, J.C.; Vorholt, J.A. The ethylmalonyl-CoA pathway is used in place of the glyoxylate cycle by Methylobacterium. J. Biol. 2012, 287, 757–766. [Google Scholar]
- Youard, Z.A.; Abicht, H.K.; Mohorko, E.; Serventi, F.; Rigozzi, E.; Ledermann, R.; Fischer, H.M.; Glockshuber, R.; Hennecke, H. A plethora of terminal oxidases and their biogenesis factors in Bradyrhizobium japonicum. In Biological Nitrogen Fixation; de Bruijn, F.J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; Volume 1, pp. 293–305. [Google Scholar]
| Strain or Plasmid | Relevant Genotype or Phenotype | Reference or Source |
|---|---|---|
| Strains | ||
| Bradyrhizobium diazoefficiens | ||
| 110spc4 | Spr wild type | [18] |
| 6806 | Spr Kmr soxR::aphII (same orientation) | This study |
| 6807 | Spr Kmr soxR::aphII (opposite orientation) | This study |
| 2494 | Spr Kmr cbbR::aphII | This study |
| 2426 | Spr Smr regR::Ω | [15] |
| cox132 | Spr Kmr coxA::Tn5 | [21] |
| 3447 | Spr Gmr cycA::gen | [22] |
| C3505 | Spr Smr cycB::Ω | [23] |
| C3524 | Spr Smr Kmr cycB::Ω cycC::aphII | [22] |
| 3448 | Spr Smr Kmr Gmr cycA::gen cycB::Ω cycC::aphII | [22] |
| Escherichia coli | ||
| BL21 (DE3) | E. coli B F− dcm ompT hsdSB (rB− mB−) gal λ(DE3) | [24] |
| DH5α | supE44 ∆lacU169 (Φ80lacZ∆M15) hsdR17 recA1 gyrA96 thi-1 relA1 | BRL a |
| S17-1 | Smr Spr hsdR (RP4-2 kan::Tn7 tet::Mu; integrated in the chromosome) | [25] |
| Plasmids | ||
| pBSL14 | Apr Kmr | [26] |
| pBSL86 | Apr Kmr | [26] |
| pSUP202pol4 | Tcr (pSUP202) oriT of RP4 | [27] |
| pUC18 | Apr cloning vector | [28] |
| pBluescript II SK+ | Apr cloning vector | Stratagene b |
| pRJ2492 | Apr (pBluescript II SK+) genomic 3,182-bp NotI-BamHI fragment spanning cbbR-cbbF’ | This study |
| pRJ2493 | Tcr (pSUP202pol4) cbbR-cbbF’, 2,353-bp EcoRI fragment of pRJ2492 | This study |
| pRJ2494 | Tcr Kmr (pSUP202pol4) ∆cbbR::aphII (same orientation), 1,211-bp Acc65I-SalI fragment of pBSL14 inserted into Acc65I-XhoI-digested pRJ2493 | This study |
| pRJ6802 | Apr (pUC18) overlapped-extension PCR-generated in frame deletion of soxR on 1.6-kb EcoRI-HindIII fragment cloned into pUC18 | This study |
| pRJ6803 | Apr Kmr (pRJ6802) ∆soxR::aphII, 1,260-bp KpnI fragment from pBSL86 inserted into KpnI site of EcoRI-HindIII fragment (same orientation as soxR) | This study |
| pRJ6804 | Apr Kmr (pRJ6802) ∆soxR::aphII, 1,260-bp KpnI fragment from pBSL86 inserted into KpnI site of EcoRI-HindIII fragment (opposite orientation to soxR) | This study |
| pRJ6806 | Kmr Tcr (pSUP202pol4) ∆soxR::aphII, 2,894-bp EcoRI/HindIII fragment from pRJ6803 inserted into SmaI site (aphII and soxR in same orientation) | This study |
| pRJ6807 | Kmr Tcr (pSUP202pol4) ∆soxR::aphII, 2,894-bp EcoRI/HindIII fragment from pRJ6804 inserted into SmaI site (aphII and soxR in opposite orientation) | This study |
| Genes | Wild Type | 6806 (ΔsoxR) |
|---|---|---|
| soxR | 16 ± 8 | --- |
| soxS | 50 ± 17 | 1.2 ± 0.5 |
| soxV | 50 ± 26 | 0.7 ± 0.3 |
| soxW | 90 ± 45 | 0.8 ± 0.7 |
| soxY1 | 4500 ± 1500 | 1.4 ± 0.4 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masuda, S.; Hennecke, H.; Fischer, H.-M. Requirements for Efficient Thiosulfate Oxidation in Bradyrhizobium diazoefficiens. Genes 2017, 8, 390. https://doi.org/10.3390/genes8120390
Masuda S, Hennecke H, Fischer H-M. Requirements for Efficient Thiosulfate Oxidation in Bradyrhizobium diazoefficiens. Genes. 2017; 8(12):390. https://doi.org/10.3390/genes8120390
Chicago/Turabian StyleMasuda, Sachiko, Hauke Hennecke, and Hans-Martin Fischer. 2017. "Requirements for Efficient Thiosulfate Oxidation in Bradyrhizobium diazoefficiens" Genes 8, no. 12: 390. https://doi.org/10.3390/genes8120390
APA StyleMasuda, S., Hennecke, H., & Fischer, H.-M. (2017). Requirements for Efficient Thiosulfate Oxidation in Bradyrhizobium diazoefficiens. Genes, 8(12), 390. https://doi.org/10.3390/genes8120390




