Transcriptome-Wide Survey and Expression Profile Analysis of Putative Chrysanthemum HD-Zip I and II Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plant Treatments
2.3. Transcriptome Search and Sequencing of Full-Length CmHB cDNAs
2.4. Phylogenetic Tree Construction and Sequence Analysis
2.5. Real-Time Quantitative PCR (qPCR)
2.6. Data Analysis
3. Results
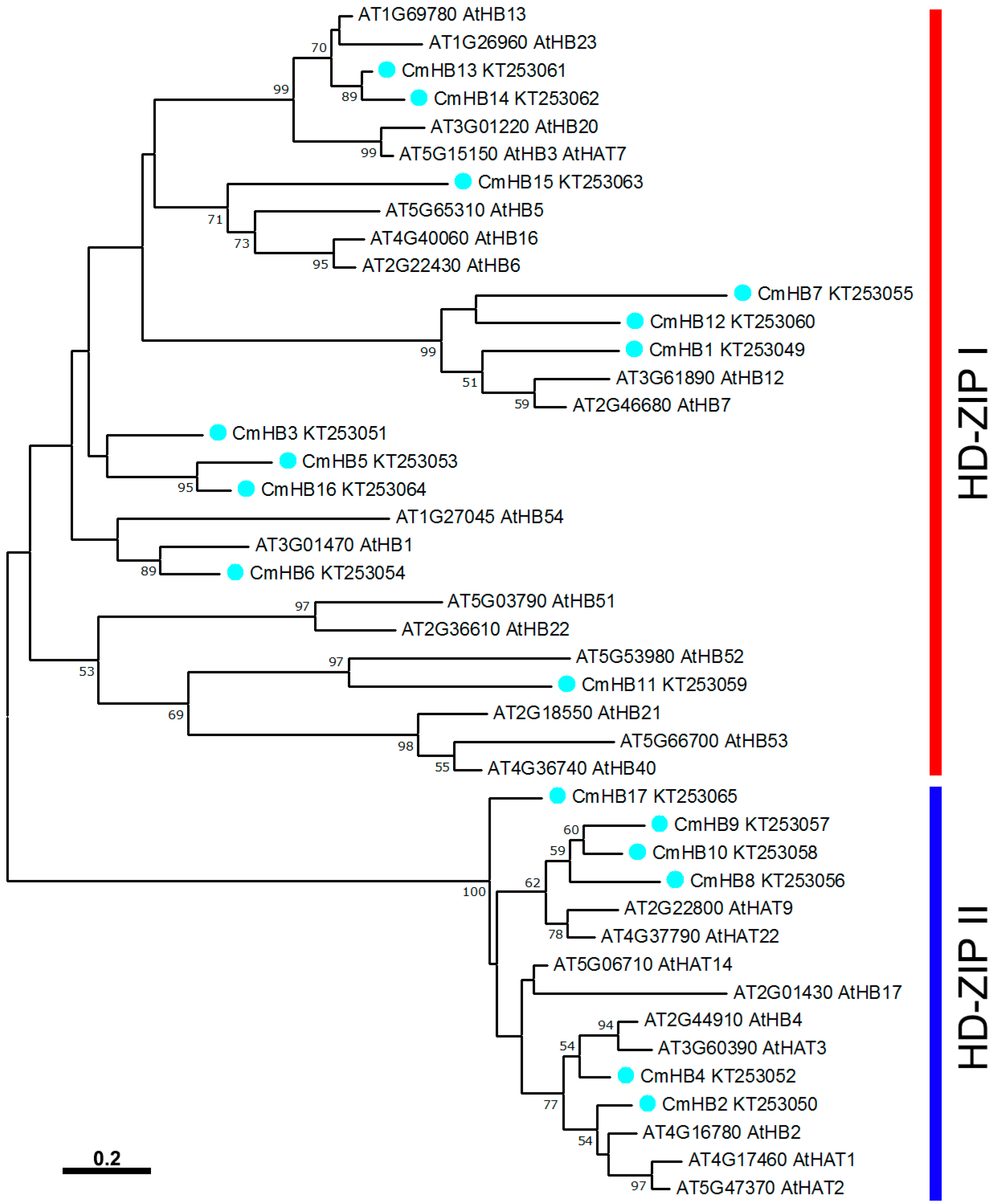
3.1. Identification and Phylogenetic Analysis of Putative HD-Zip Factors in Chrysanthemum
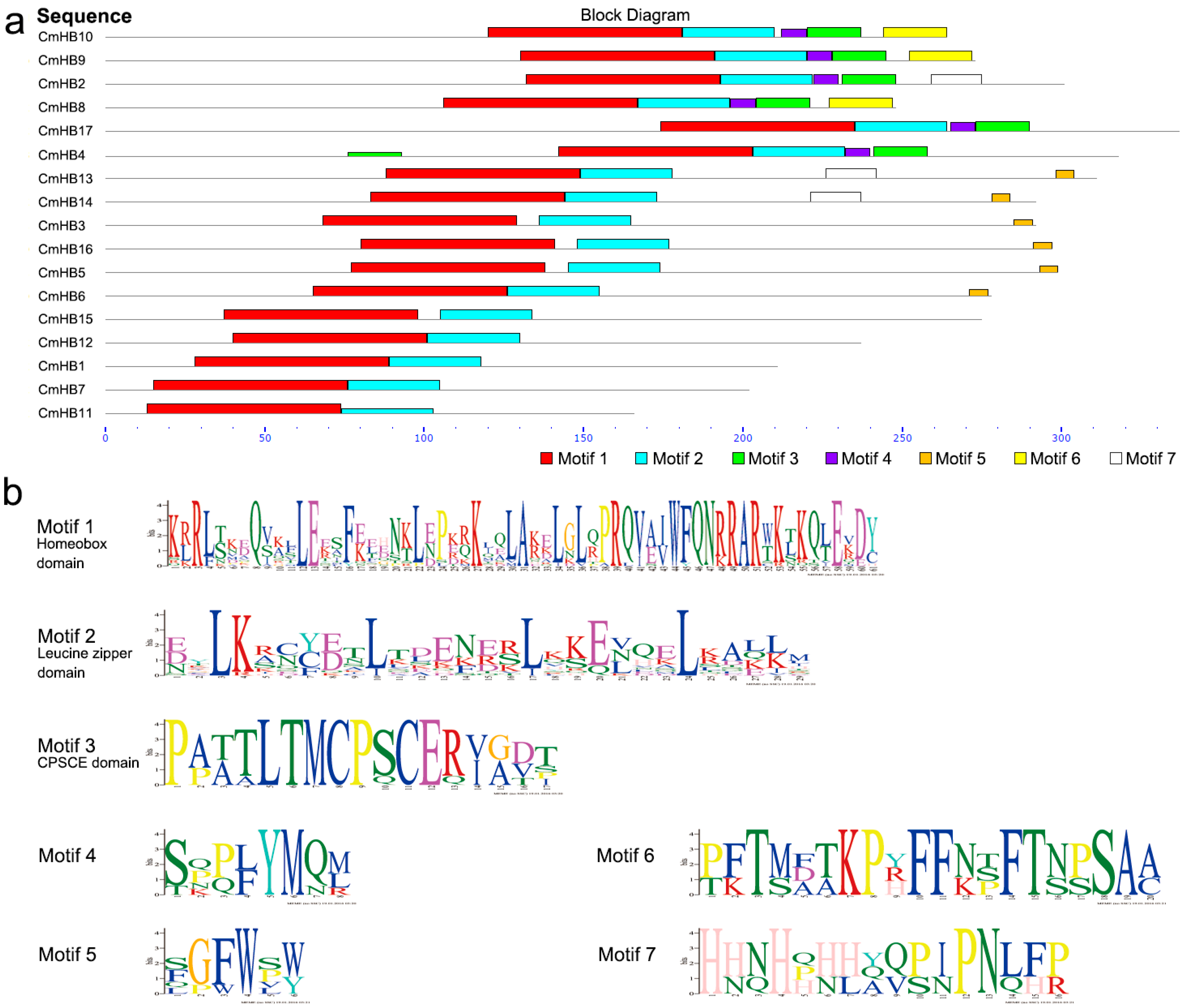
3.2. Conserved Sequences in Chrysanthemum HD-Zip Proteins
3.3. miRNA Target Site Prediction
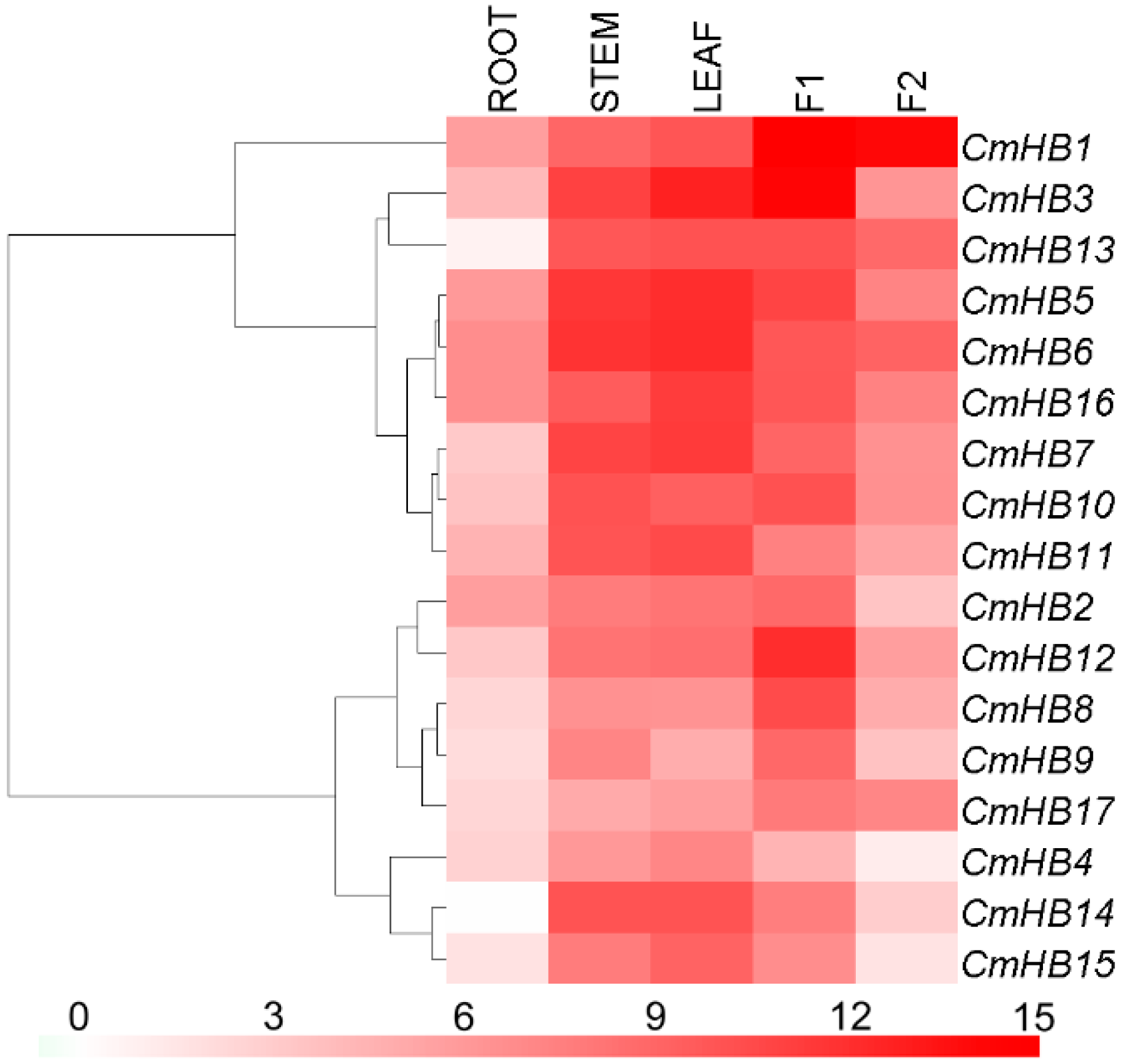
3.4. Transcription Profiling of CmHB Genes
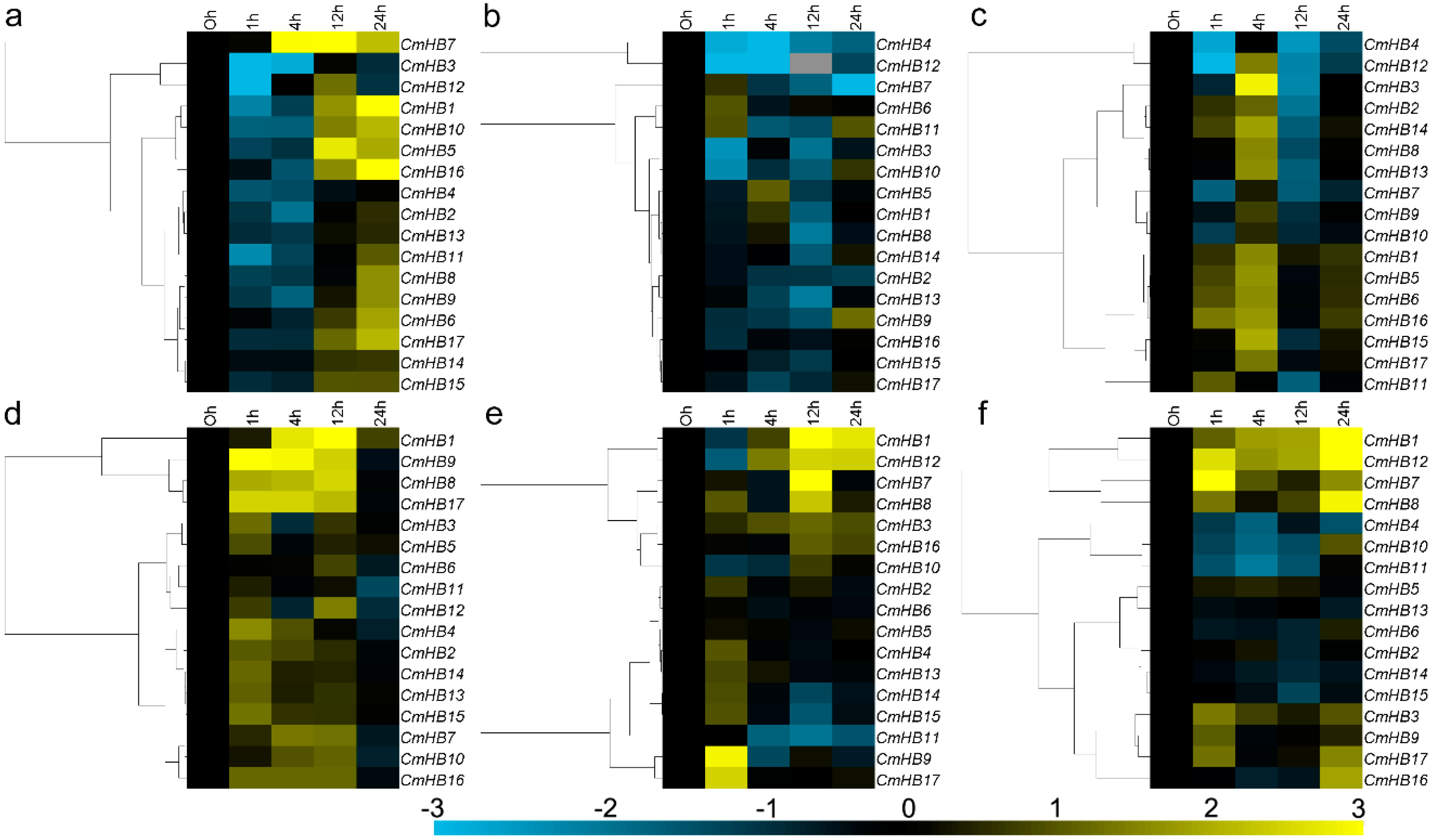
3.5. Expression of CmHB Genes After Treatment with Phytohormones
3.6. Expression Profiling of CmHB Genes Under Abiotic Stress
4. Discussion
4.1. Comparative Analysis of the Chrysanthemum and Arabidopsis HD-Zip Gene Families
4.2. miRNA Target Site Prediction
4.3. Expression Patterns of CmHB Genes
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Zalewski, C.S.; Floyd, S.K.; Furumizu, C.; Sakakibara, K.; Stevenson, D.W.; Bowman, J.L. Evolution of the class IV HD-zip gene family in streptophytes. Mol. Biol. Evol. 2013, 30, 2347–2365. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Davis, R.W. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. USA 1992, 89, 3894–3898. [Google Scholar] [CrossRef] [PubMed]
- Elhiti, M.; Stasolla, C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009, 4, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Palena, C.M.; Tron, A.E.; Bertoncini, C.W.; Gonzalez, D.H.; Chan, R.L. Positively charged residues at the N-terminal arm of the homeodomain are required for efficient DNA binding by homeodomain-leucine zipper proteins. J. Mol. Biol. 2001, 308, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Tron, A.E.; Comelli, R.N.; Gonzalez, D.H. Structure of homeodomain-leucine zipper/DNA complexes studied using hydroxyl radical cleavage of DNA and methylation interference. Biochemistry 2005, 44, 16796–16803. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Steindler, C.; Morelli, G.; Ruberti, I. The Arabidopsis Athb-8, -9 and genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 1998, 38, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Katsumata, H.; Komeda, Y.; Takahashi, T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 2003, 130, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Ciarbelli, A.R.; Ciolfi, A.; Salvucci, S.; Ruzza, V.; Possenti, M.; Carabelli, M.; Fruscalzo, A.; Sessa, G.; Morelli, G.; Ruberti, I. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol. Biol. 2008, 68, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Agalou, A.; Purwantomo, S.; Övernäs, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engström, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2008, 66, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhou, Y.; Jiang, H.; Li, X.; Gan, D.; Peng, X.; Zhu, S.; Cheng, B. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE 2011, 6, e28488. [Google Scholar] [CrossRef] [PubMed]
- Re, D.A.; Dezar, C.A.; Chan, R.L.; Baldwin, I.T.; Bonaventure, G. Nicotiana attenuata NaHD20 plays a role in leaf ABA accumulation during water stress, benzylacetone emission from flowers, and the timing of bolting and flower transitions. J. Exp. Bot. 2011, 62, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, X.; Guan, X.; Liu, Y.; Chen, H.; Wang, T.; Mouekouba, L.D.; Li, J.; Wang, A. A genome-wide survey of homeodomain-leucine zipper genes and analysis of cold-responsive HD-Zip I members’ expression in tomato. Biosci. Biotechnol. Biochem. 2014, 78, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Morishige, H.; Tanaka, T.; Kanamori, H.; Komatsuda, T.; Sato, K.; Itoh, T.; Wu, J.; Nakamura, S. Transcriptome analysis of barley identifies heat shock and HD-Zip I transcription factors up-regulated in response to multiple abiotic stresses. Mol. Breeding 2014, 34, 761–768. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Z.; Zhao, H.; Zhao, Y.; Cheng, B.; Xiang, Y. Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 2014, 9, e87156. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Chen, Y.; Zhao, M.; Zhang, W.H. A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses. Environ. Exp. Bot. 2012, 80, 1–9. [Google Scholar] [CrossRef]
- Rueda, E.; Dezar, C.; Gonzalez, D.; Chan, R. Hahb-10, a sunflower homeobox-leucine zipper gene, is involved in the response to dark/light conditions and promotes a reduction of the life cycle when expressed in Arabidopsis. Plant Cell Physiol. 2005, 46, 1954–1963. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, K.; Nishiyama, T.; Kato, M.; Hasebe, M. Isolation of homeodomain–leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain–leucine zipper genes in land plants. Mol. Biol. Evol. 2001, 18, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Chi, X.; Chai, G.; Kong, Y.; He, G.; Wang, X.; Shi, D.; Zhang, D.; Zhou, G. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PloS ONE 2012, 7, e31149. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, E.; Olsson, A.S.; Johannesson, H.; Johansson, H.; Hanson, J.; Engström, P.; Söderman, E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Dong, C.H.; Wu, Y.; Carabelli, M.; Sessa, G.; Ruberti, I.; Morelli, G.; Chua, N.H. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell 1995, 7, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, Q.; Jin, X.; Peng, X.; Liu, J.; Deng, L.; Yan, H.; Sheng, L.; Jiang, H.; Cheng, B. A novel maize homeodomain–leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and arabidopsis. Plant Cell Physiol. 2014, 55, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- De Smet, I.; Lau, S.; Ehrismann, J.S.; Axiotis, I.; Kolb, M.; Kientz, M.; Weijers, D.; Jürgens, G. Transcriptional repression of BODENLOS by HD-ZIP transcription factor HB5 in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 3009–3019. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, S.; Pourkheirandish, M.; Hensel, G.; Kumlehn, J.; Stein, N.; Tagiri, A.; Yamaji, N.; Ma, J.F.; Sassa, H.; Koba, T. Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol. 2013, 197, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Turchi, L.; Baima, S.; Morelli, G.; Ruberti, I. Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 2015, 66, 5043–5053. [Google Scholar] [CrossRef] [PubMed]
- Sawa, S.; Ohgishi, M.; Goda, H.; Higuchi, K.; Shimada, Y.; Yoshida, S.; Koshiba, T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 2002, 32, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Reymond, M.C.; Brunoud, G.; Chauvet, A.; Martínez-Garcia, J.F.; Martin-Magniette, M.L.; Monéger, F.; Scutt, C.P. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 2012, 24, 2812–2825. [Google Scholar] [CrossRef] [PubMed]
- Bou-Torrent, J.; Salla-Martret, M.; Brandt, R.; Musielak, T.; Palauqui, J.C.; Martínez-García, J.F.; Wenkel, S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 2012, 7, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Turchi, L.; Carabelli, M.; Ruzza, V.; Possenti, M.; Sassi, M.; Peñalosa, A.; Sessa, G.; Salvi, S.; Forte, V.; Morelli, G. Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 2013, 140, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.; Roberts, C.; Carlsbecker, A. HD-Zip class III transcription factors control root development through the modulation of ROS levels. Biotechnologia 2013, 94, 217–229. [Google Scholar]
- Côté, C.L.; Boileau, F.; Roy, V.; Ouellet, M.; Levasseur, C.; Morency, M.J.; Cooke, J.E.; Séguin, A.; MacKay, J.J. Gene family structure, expression and functional analysis of HD-Zip III genes in angiosperm and gymnosperm forest trees. BMC Plant Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Baima, S.; Possenti, M.; Matteucci, A.; Wisman, E.; Altamura, M.M.; Ruberti, I.; Morelli, G. The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001, 126, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Robischon, M.; Du, J.; Miura, E.; Groover, A. The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 2011, 155, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Vernoud, V.; Laigle, G.; Rozier, F.; Meeley, R.B.; Perez, P.; Rogowsky, P.M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009, 59, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, X.; Wang, Z.; Wang, S.; Wang, Y.; Zhu, Q.; Li, S.; Xiang, C. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013, 162, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Kamata, N.; Okada, H.; Komeda, Y.; Takahashi, T. Mutations in epidermis-specific HD-ZIP IV genes affect floral organ identity in Arabidopsis thaliana. Plant J. 2013, 75, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, E.; Yamada, Y.; Sezaki, N.; Kosaka, S.; Kondo, H.; Kamata, N.; Abe, M.; Komeda, Y.; Takahashi, T. ATML1 and PDF2 play a redundant and essential role in Arabidopsis embryo development. Plant Cell Physiol. 2015, 56, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Khosla, A.; Boehler, A.P.; Bradley, A.M.; Neumann, T.R.; Schrick, K. HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 2014, 26, 2184–2200. [Google Scholar] [CrossRef] [PubMed]
- Dezar, C.A.; Giacomelli, J.I.; Manavella, P.A.; Ré, D.A.; Alves-Ferreira, M.; Baldwin, I.T.; Bonaventure, G.; Chan, R.L. HAHB10, a sunflower HD-Zip II transcription factor, participates in the induction of flowering and in the control of phytohormone-mediated responses to biotic stress. J. Exp. Bot. 2011, 62, 1061–1076. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lin, J.; Li, X.G.; Chang, Y. Genome-wide identification of pear HD-Zip gene family and expression patterns under stress induced by drought, salinity, and pathogen. Acta Physiol. Plant 2015, 37, 1–19. [Google Scholar] [CrossRef]
- Manavella, P.A.; Dezar, C.A.; Bonaventure, G.; Baldwin, I.T.; Chan, R.L. HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 2008, 56, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ma, R.; Shen, Z.; Sun, X.; Korir, N.; Yu, M. Genome-wide analysis of the homeodomain-leucine zipper (HD-ZIP) gene family in peach (Prunus persica). Genet. Mol. Res. 2014, 13, 2654–2668. [Google Scholar] [CrossRef] [PubMed]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Song, A.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol. Biol. Rep. 2014, 41, 4155–4162. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Lu, J.; Jiang, J.; Chen, S.; Guan, Z.; Fang, W.; Chen, F. Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na+/H+ antiporter. Plant Biol. 2012, 14, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; An, J.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 2014, 80, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Li, P.; Jiang, J.; Chen, S.; Li, H.; Zeng, J.; Shao, Y.; Zhu, L.; Zhang, Z.; Chen, F. Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 2014, 15, 14442–14455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Z.; Dong, W.; Sun, C.; Wang, H.; Song, A.; He, L.; Fang, W.; Chen, F.; Teng, N. Transcriptomic and proteomic analysis reveals mechanisms of embryo abortion during chrysanthemum cross breeding. Sci. Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- de Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Wu, D.; Fan, Q.; Tian, C.; Chen, S.; Guan, Z.; Xin, J.; Zhao, K.; Chen, F. Transcriptome-wide identification and expression profiling analysis of chrysanthemum trihelix transcription factors. Int. J. Mol. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Schliep, M.; Ebert, B.; Simon-Rosin, U.; Zoeller, D.; Fisahn, J. Quantitative expression analysis of selected transcription factors in pavement, basal and trichome cells of mature leaves from Arabidopsis thaliana. Protoplasma 2010, 241, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Morelli, G.; Baima, S.; Carabelli, M.; di Cristina, M.; Lucchetti, S.; Sessa, G.; Steindler, C.; Ruberti, I. Homeodomain-leucine zipper proteins in the control of plant growth and development. In Cellular Integration of Signalling Pathways in Plant Development; Springer: Berlin, Germany, 1998; pp. 251–262. [Google Scholar]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.C.; Hrmova, M.; Lopato, S.; Langridge, P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011, 190, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Blumwald, E. The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 2015, 27, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.E.; Övernäs, E.; Johansson, H.; Rada-Iglesias, A.; Engström, P. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 2012, 80, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Söderman, E.; Hjellström, M.; Fahleson, J.; Engström, P. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol. Biol. 1999, 40, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Vos, I.A.; Moritz, L.; Pieterse, C.M.; Van Wees, S.C. Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front Plant Sci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.S.; Engström, P.; Söderman, E. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004, 55, 663–677. [Google Scholar] [CrossRef] [PubMed]
| Gene | GenBank Accession No. | Amino Acid Length (aa) | PI | MW | Subcellular Localization | Most Similar A. thaliana Homolog | Locus Name |
|---|---|---|---|---|---|---|---|
| CmHB1 | KT253049 | 211 | 4.98 | 24656.44 | N(9.05) | ATHB7 | AT2G46680.1 |
| CmHB2 | KT253050 | 301 | 8.82 | 33945.17 | N(9.30) | ATHB2 | AT4G16780.1 |
| CmHB3 | KT253051 | 292 | 4.85 | 34011.51 | N(7.93) | ATHB1 | AT3G01470.1 |
| CmHB4 | KT253052 | 318 | 8.7 | 35070.59 | N(9.40) | HAT3 | AT3G60390.1 |
| CmHB5 | KT253053 | 299 | 4.71 | 33985.07 | N(7.97) | ATHB1 | AT3G01470.1 |
| CmHB6 | KT253054 | 278 | 4.66 | 31919.69 | N(8.94) | ATHB1 | AT3G01470.1 |
| CmHB7 | KT253055 | 202 | 6.61 | 23500.1 | N(8.95) | ATHB7 | AT2G46680.1 |
| CmHB8 | KT253056 | 248 | 8.6 | 27814.11 | N(9.47) | HAT22 | AT4G37790.1 |
| CmHB9 | KT253057 | 273 | 8.46 | 30384.22 | N(9.32) | HAT22 | AT4G37790.1 |
| CmHB10 | KT253058 | 264 | 9.13 | 29681.66 | N(9.36) | HAT9 | AT2G22800.1 |
| CmHB11 | KT253059 | 166 | 8.85 | 19298.5 | N(8.76) | ATHB52 | AT5G53980.1 |
| CmHB12 | KT253060 | 237 | 5.45 | 27259.88 | N(8.66) | ATHB12 | AT3G61890.1 |
| CmHB13 | KT253061 | 311 | 5.91 | 35462.34 | N(9.18) | ATHB13 | AT1G69780.1 |
| CmHB14 | KT253062 | 292 | 5.76 | 33627.57 | N(8.85) | ATHB13 | AT1G69780.1 |
| CmHB15 | KT253063 | 275 | 4.69 | 31595.82 | N(8.68) | ATHB6 | AT2G22430.1 |
| CmHB16 | KT253064 | 297 | 4.91 | 34061.3 | N(7.97) | ATHB1 | AT3G01470.1 |
| CmHB17 | KT253065 | 337 | 7.63 | 37486.68 | N(9.08) | HAT14 | AT5G06710.1 |
| miRNA | Target | Exp | UPE | miRNA Start | miRNA End | Target Start | Target End |
|---|---|---|---|---|---|---|---|
| miR414 | CmHB4 | 2.5 | 20.266 | 1 | 21 | 512 | 532 |
| miR414 | CmHB9 | 2.5 | 10.125 | 1 | 21 | 460 | 480 |
| miR917 | CmHB9 | 2.5 | 18.596 | 1 | 20 | 360 | 379 |
| miR1023a-3p | CmHB13 | 3 | 13.468 | 1 | 20 | 135 | 154 |
| miR444 | CmHB16 | 2 | 9.773 | 1 | 21 | 537 | 557 |
| miR158a-5p | CmHB5 | 3 | 19.317 | 1 | 19 | 738 | 757 |
| miR4369 | CmHB17 | 3 | 16.17 | 1 | 20 | 368 | 387 |
| miR4993 | CmHB4 | 3 | 17.178 | 1 | 21 | 922 | 942 |
| miR5298 | CmHB10 | 3 | 5.976 | 1 | 22 | 135 | 156 |
| miR5380 | CmHB9 | 3 | 1.732 | 1 | 23 | 9 | 31 |
| miR5751 | CmHB14 | 3 | 16.3 | 1 | 20 | 134 | 153 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, A.; Li, P.; Xin, J.; Chen, S.; Zhao, K.; Wu, D.; Fan, Q.; Gao, T.; Chen, F.; Guan, Z. Transcriptome-Wide Survey and Expression Profile Analysis of Putative Chrysanthemum HD-Zip I and II Genes. Genes 2016, 7, 19. https://doi.org/10.3390/genes7050019
Song A, Li P, Xin J, Chen S, Zhao K, Wu D, Fan Q, Gao T, Chen F, Guan Z. Transcriptome-Wide Survey and Expression Profile Analysis of Putative Chrysanthemum HD-Zip I and II Genes. Genes. 2016; 7(5):19. https://doi.org/10.3390/genes7050019
Chicago/Turabian StyleSong, Aiping, Peiling Li, Jingjing Xin, Sumei Chen, Kunkun Zhao, Dan Wu, Qingqing Fan, Tianwei Gao, Fadi Chen, and Zhiyong Guan. 2016. "Transcriptome-Wide Survey and Expression Profile Analysis of Putative Chrysanthemum HD-Zip I and II Genes" Genes 7, no. 5: 19. https://doi.org/10.3390/genes7050019
APA StyleSong, A., Li, P., Xin, J., Chen, S., Zhao, K., Wu, D., Fan, Q., Gao, T., Chen, F., & Guan, Z. (2016). Transcriptome-Wide Survey and Expression Profile Analysis of Putative Chrysanthemum HD-Zip I and II Genes. Genes, 7(5), 19. https://doi.org/10.3390/genes7050019








