Matricellular Signal Transduction Involving Calmodulin in the Social Amoebozoan Dictyostelium
Abstract
:1. Introduction
1.1. Matricellular Protein-Mediated Signal Transduction
1.2. ECM Proteins of Dictyostelium discoideum
2. Current Research
2.1. CyrA is a CaM-binding EGFL Repeat-Containing ECM Protein
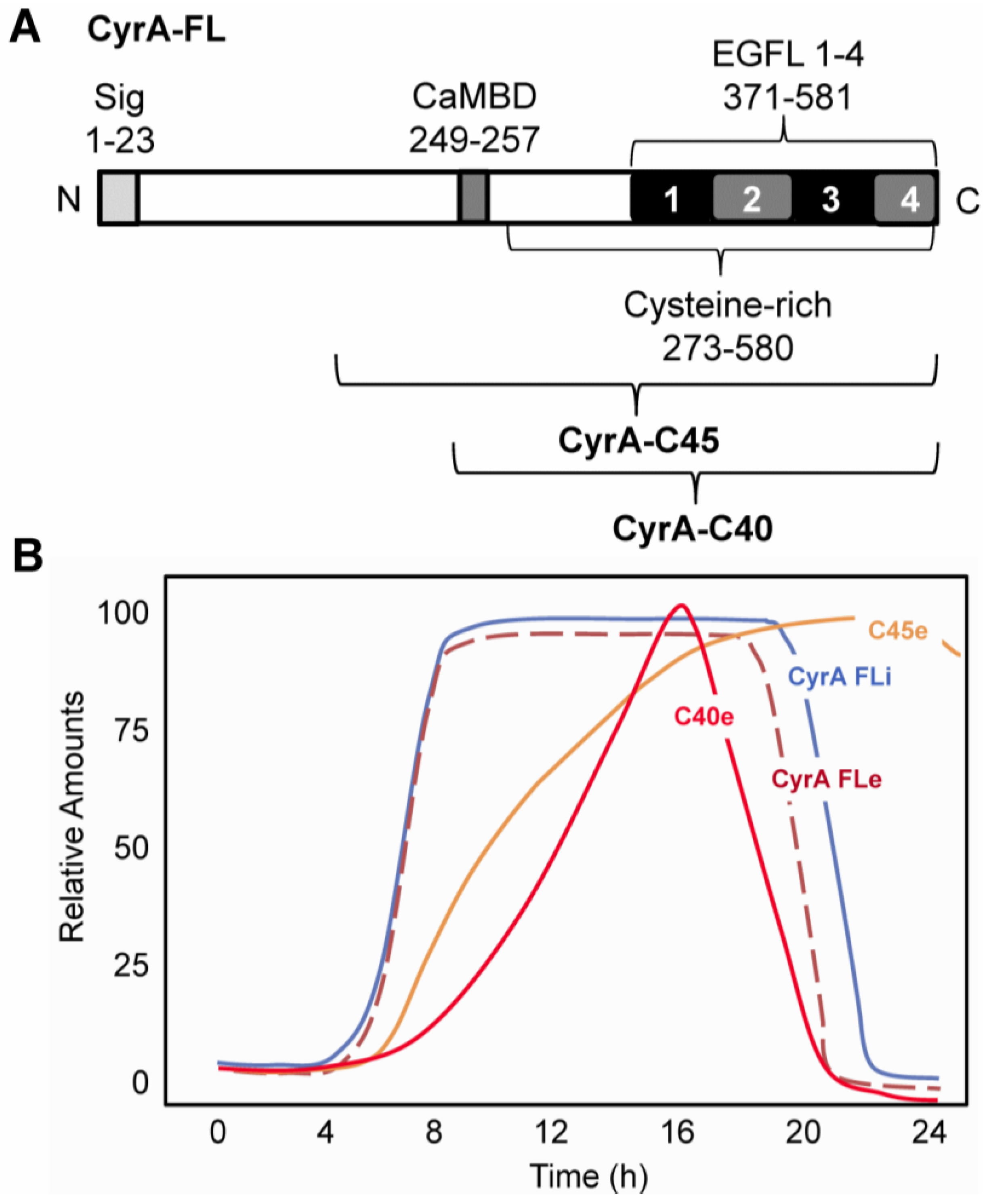
2.2. EGFL1 Peptide Increases Cell Motility and Chemotaxis
2.3. EGFL1-Mediated Signal Transduction
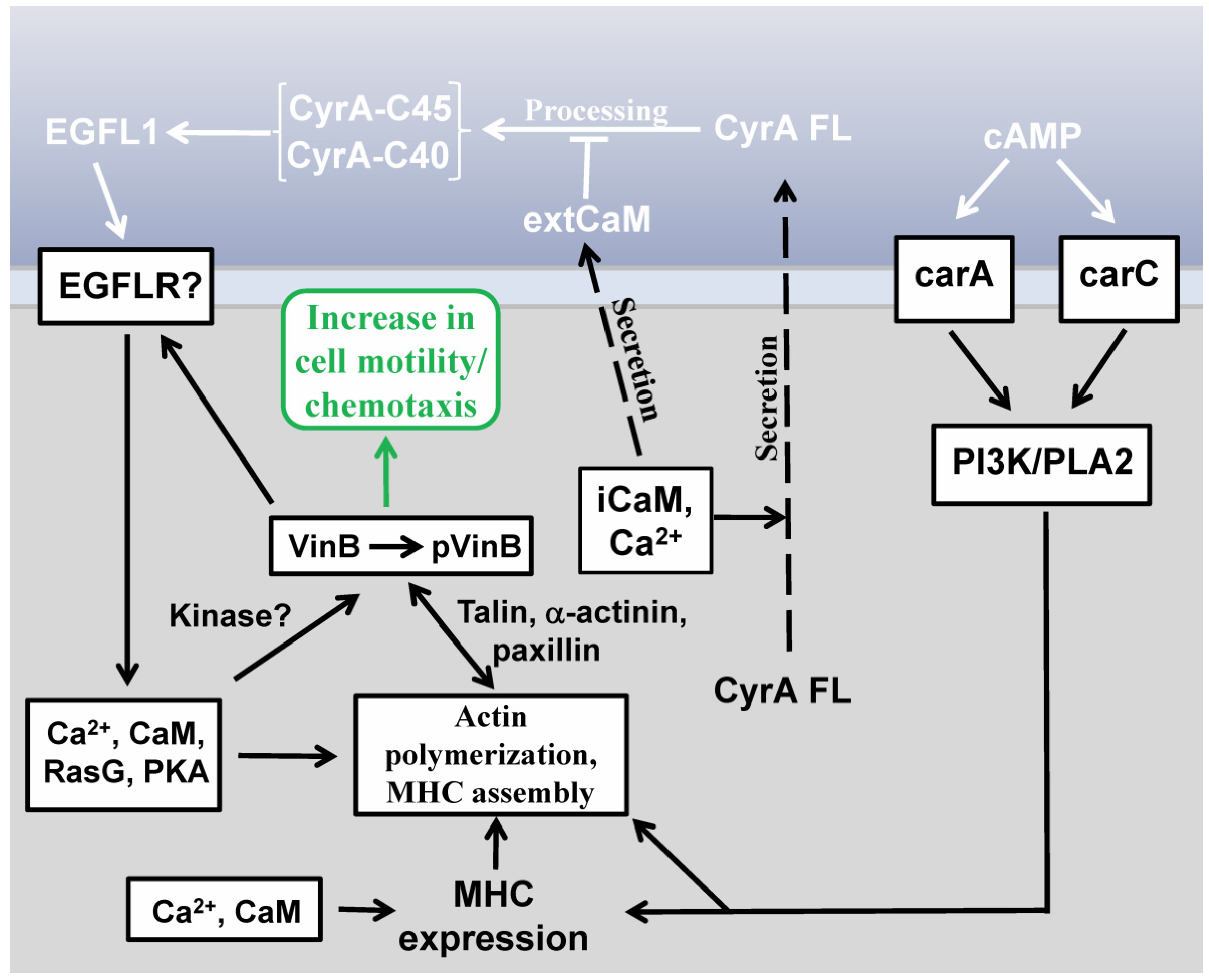
2.4. Vin B Phosphorylation is Regulated by EGFL1 Signal Transduction
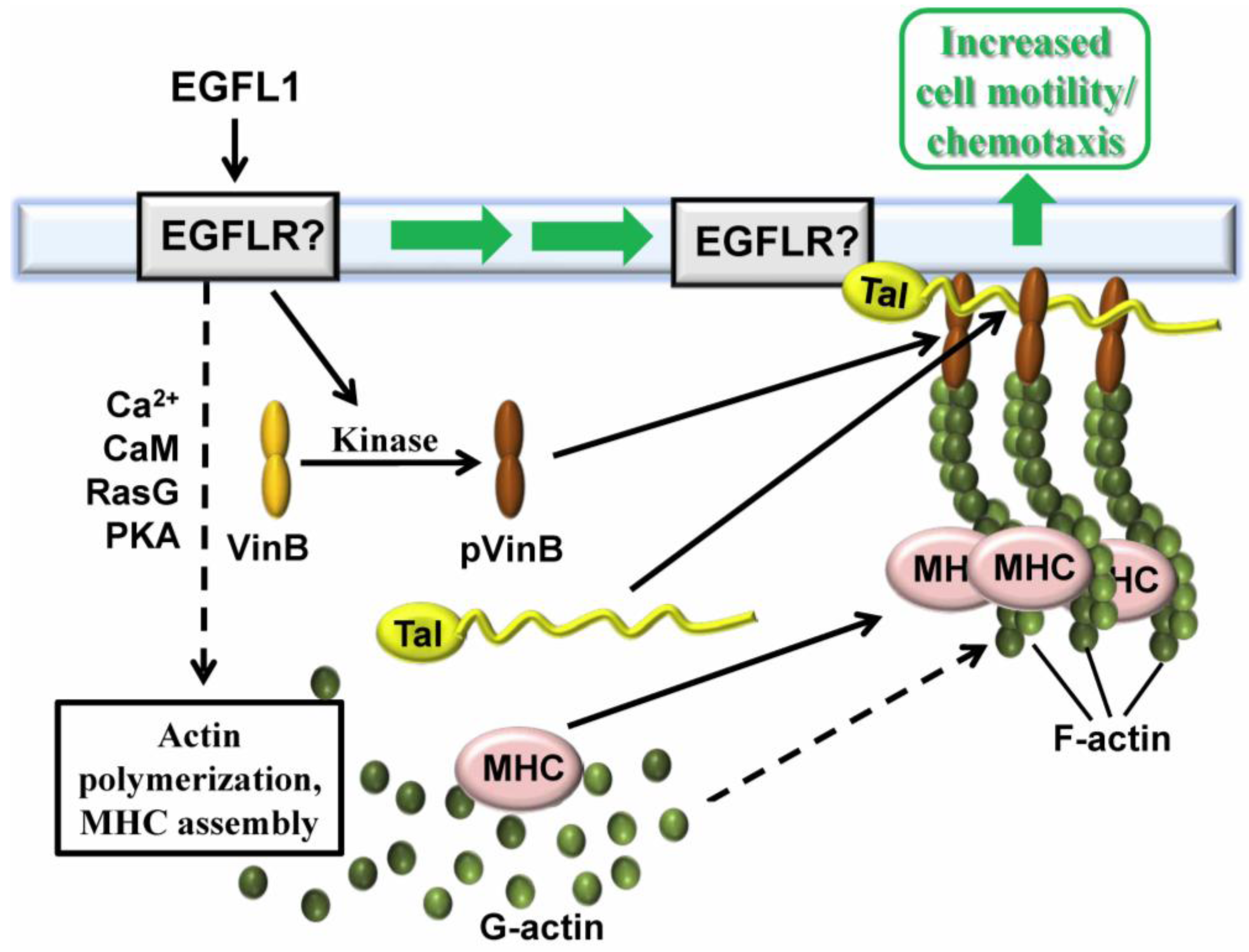
2.5. Extracellular CaM and its Multiple Functions
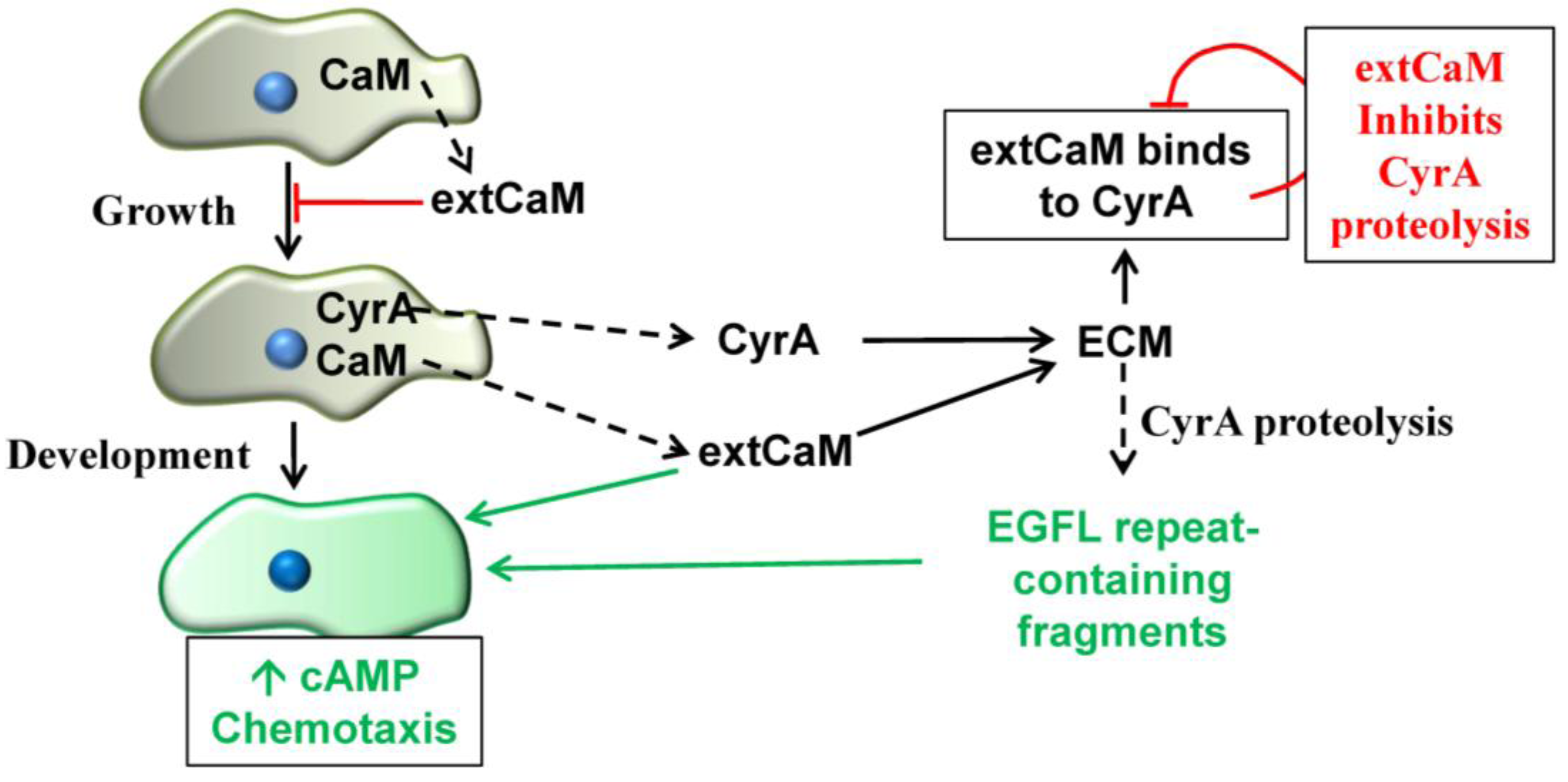
3. Conclusions
Acknowledgements
References
- Bornstein, P.; Sage, E.H. Matricellular proteins: extracellular modulators of cell function. Curr. Opin. Cell. Biol. 2002, 14, 608–616. [Google Scholar] [CrossRef]
- Roberts, D.D. Emerging functions of matricellular proteins. Cell. Mol. Life. Sci. 2011, 68, 3133–3136. [Google Scholar] [CrossRef]
- Mosher, D.F.; Adams, J.C. Adhesion-modulating/matricellular ECM protein families: A structural, functional and evolutionary appraisal. Matrix Biol. 2012, 31, 155–161. [Google Scholar] [CrossRef]
- Adams, J.C.; Lawler, J. The thrombospondins. Cold Spring Harb. Perspect. Biol. 2011, 3, a009712. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Diverse biological functions of the SPARC family of proteins. Int. J. Biochem. Cell Biol. 2012, 44, 480–488. [Google Scholar] [CrossRef]
- Apte, S.S. A disintegrin-like and metalloproteinase (reprolysin-type) with thrombospondin type I motif (ADAMTS) superfamily – functions and mechanisms. J. Biol. Chem. 2009, 284, 31493–31497. [Google Scholar] [CrossRef]
- Campbell, I.; Bork, P. Epidermal growth factor-like modules. Curr. Opin. Struct. Biol. 1993, 3, 385–392. [Google Scholar] [CrossRef]
- Rao, Z.; Handford, P.; Mayhew, M.; Knott, V.; Browniee, G.C.; Stuart, D. The structure of a Ca2+-binding epidermal growth factor-like domain: Its role in protein-protein interactions. Cell 1995, 82, 131–141. [Google Scholar] [CrossRef]
- Fey, P.; Stephens, S.; Titus, M.A.; Chisholm, R.L. SadA, a novel adhesion receptor in Dictyostelium. J. Cell Biol. 2002, 159, 1109–1119. [Google Scholar] [CrossRef]
- Glöckner, G.; Eichinger, L.; Szafranski, K.; Pachebat, J.A.; Bankier, A.T.; Dear, P.H.; Lehmann, R.; Baumgart, C.; Parra, G.; Abril, J.F.; Guigó, R.; Kumpf, K.; Tunggal, B.; Cox, E.; Quail, M.A.; Platzer, M.; Rosenthal, A.; Noegel, A.A. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 2002, 418, 79–85. [Google Scholar]
- Kurucz, E.; Márkus, R.; Zsámboki, J.; Folkl-Medzihradszky, K.; Darula, Z.; Vilmos, P.; Udvardy, A.; Krausz, I.; Lukacsovich, T.; Gateff, E.; Zettervall, C.J.; Hultmark, D.; Andó, I. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 2007, 17, 649–654. [Google Scholar]
- Iyer, A.K.; Kien, T.T.; Borysenko, C.W.; Cascio, M.; Camacho, C.J.; Blair, H.C.; Bahar, I.; Wells, A. Tenascin cytotactin epidermal growth factor-like repeat binds epidermal growth factor receptor with low affinity. J. Cell Physiol. 2007, 211, 748–758. [Google Scholar] [CrossRef]
- Iyer, A.K.V.; Tran, K.T.; Griffith, L.; Wells, A. Cell surface restriction of EGFR by a tenascin cytotactin-encoded EGF-like repeat is preferential for motility-related signalling. J. Cell Physiol. 2008, 214, 504–512. [Google Scholar] [CrossRef]
- Swindle, C.S.; Tran, K.T.; Johnson, T.D.; Banerjee, P.; Mayes, A.M.; Griffith, L.; Wells, A. Epidermal growth factor (EGF)-like repeats of human Tenascin-C as ligands for EGF receptor. J. Cell Biol. 2001, 154, 459–468. [Google Scholar] [CrossRef]
- Liu, A.; Garg, P.; Yang, S.; Gong, P.; Pallero, M.A.; Annis, D.S.; Liu, Y.; Passaniti, A.; Mann, D.; Mosher, D.F.; Murphy-Ullrich, J.E.; Goldblum, S.E. Epidermal growth factor-like repeats of thrombospondins activate phospholipase Cgamma and increase epithelial cell migration through indirect epidermal growth factor receptor activation. J. Biol. Chem. 2009, 284, 6389–6402. [Google Scholar]
- Schenk, S.; Hintermann, E.; Bilban, M.; Koshikawa, N.; Hojilla, C.; Khokha, R.; Quaranta, V. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMP-dependent mammary gland involution. J. Cell. Biol. 2003, 161, 197–209. [Google Scholar] [CrossRef]
- Williams, J.G. Dictyostelium finds new roles to model. Genetics 2010, 185, 717–726. [Google Scholar] [CrossRef]
- Insall, R.; Andrew, N. Chemotaxis in Dictyostelium: how to walk straight using parallel pathways. Curr. Opin. Microbiol. 2007, 10, 578–581. [Google Scholar] [CrossRef]
- Jin, T.; Hereld, D. Moving toward understanding eukaryotic chemotaxis. Eur. J. Cell Biol. 2006, 85, 905–913. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, C-L.; Iijima, M. Signaling mechanisms for chemotaxis. Dev. Growth Differen. 2011, 53, 495–502. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Williams, K.L. The extracellular matrix of the Dictyostelium discoideum slug. Experientia 1995, 51, 1189–1196. [Google Scholar] [CrossRef]
- Schaap, P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum. Development 2011, 138, 387–396. [Google Scholar] [CrossRef]
- Breen, E.J.; Vardy, P.H.; Williams, K.L. Movement of the multicellular slug stage of Dictyostelium discoideum: an analytical approach. Development 1987, 101, 313–321. [Google Scholar]
- Grant, W.N.; Williams, K.L. Monoclonal antibody characterization of the slime sheath: The extracellular matrix of Dictyostelium discoideum. EMBO J. 1983, 2, 935–940. [Google Scholar]
- Morrison, A.; Blanton, R.L.; Grimson, M.; Fuchs, M.; Williams, K.L.; Williams, J. Disruption of the gene encoding the EcmA, extracellular matrix protein of Dictyostelium, alters slug morphology. Dev. Biol. 1994, 163, 457–466. [Google Scholar] [CrossRef]
- Breen, E.J.; Williams, K.L. Movement of the Dictyostelium discoideum slug: models, musings and images. Dev. Genet. 1988, 9, 539–548. [Google Scholar] [CrossRef]
- Suarez, A.; Huber, R.J.; Myre, M.A.; O’Day, D.H. An extracellular matrix, calmodulin-binding protein from Dictyostelium with EGF-like repeats that enhance cell motility. Cell. Signal. 2011, 23, 1197–1206. [Google Scholar] [CrossRef]
- Huber, R.J.; Suarez, A.; O’Day, D.H. CyrA, a matricellular protein that modulates cell motility in Dictyostelium discoideum. Matrix Biol. 2012, 31, 271–280. [Google Scholar] [CrossRef]
- O’Day, D. H. CaMBOT: Profiling and Characterizing Calmodulin Binding Proteins. Cell. Signal. 2003, 15, 347–355. [Google Scholar] [CrossRef]
- Bakthavatsalam, D.; Gomer, R.H. The secreted proteome profile of developing Dictyostelium discoideum cells. Proteomics 2010, 10, 2556–2559. [Google Scholar] [CrossRef]
- Huber, R.J.; O’Day, D.H. An EGF-like peptide sequence from Dictyostelium enhances cell motility and chemotaxis. Biochem. Biophys. Res. Commun. 2009, 379, 470–475. [Google Scholar] [CrossRef]
- Huber, R.J.; O’Day, D.H. EGF-like peptide-enhanced cell motility in Dictyostelium functions independently of the cAMP-mediated pathway and requires active Ca2+/calmodulin signaling. Cell. Signal. 2011, 23, 731–738. [Google Scholar] [CrossRef]
- Nikolaeva, I.; Huber, R.J.; O’Day, D.H. EGF-like peptide of Dictyostelium discoideum is not a chemoattractant but it does restore folate-mediated chemotaxis in the presence of signal transduction inhibitors. Peptides 2012, 34, 145–149. [Google Scholar] [CrossRef]
- Huber, R.J.; O’Day, D.H. A Matricellular protein and EGF-like repeat signalling in the social amoebozoan Dictyostelium discoideum. Cell. Mol. Life Sci. 2012, 69, 3989–3997. [Google Scholar] [CrossRef]
- Huber, R.J.; O’Day, D.H. EGF-like peptide-enhanced cell movement in Dictyostelium is mediated by protein kinases and the activity of several cytoskeletal proteins. Cell. Signal. 2012, 24, 1770–1780. [Google Scholar] [CrossRef]
- Bryant, J.A.; Finn, R.S.; Slamon, D.J.; Cloughesy, T.F.; Charles, A.C. EGF activates intracellular and intercellular calcium signaling by distinct pathways in tumor cells. Cancer Biol. Ther. 2004, 3, 1243–1249. [Google Scholar] [CrossRef]
- Kato, K.; Ueoka, Y.; Tamura, T.; Nishida, J.; Wake, N. Oncogenic Ras modulates epidermal growth factor responsiveness in endometrial carcinomas. Eur. J. Cancer 1998, 34, 737–744. [Google Scholar] [CrossRef]
- Zhang, H.; Heid, P.J.; Wessels, D.; Daniels, K.J.; Pham, T.; Loomis, W.F.; Soll, D.R. Constitutively active protein kinase A disrupts motility and chemotaxis in Dictyostelium discoideum. Eukaryot. Cell. 2003, 2, 62–75. [Google Scholar] [CrossRef]
- Sasaki, A.T.; Janetopoulos, C.; Lee, S.; Charest, P.G.; Takeda, K.; Sundheimer, L.W.; Meili, R.; Devreotes, P.N.; Firtel, R.A. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 2007, 178, 185–191. [Google Scholar] [CrossRef]
- Wu, L.; Valkema, R.; Van Haastert, P.J.; Devreotes, P.N. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J. Cell Biol. 1995, 129, 1667–1675. [Google Scholar] [CrossRef]
- Sun, W.; Li, S.; Xu, J.; Liu, T.; Shang, Z. H+-ATPase in the plasma membrane of Arabidopsis pollen cells is involved in extracellular calmodulin-promoted pollen germination. Prog. Nat. Sci. 2009, 19, 1071–1078. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, Y.-Q.; Lü, Y.-Q.; Lin, H.F.; Bai, Y.; Wang, X.C.; Chen, Y.L. A signaling pathway linking nitric oxide production to heterotrimeric G protein and hydrogen peroxide regulates extracellular calmodulin induction of stomatal closure in Arabidopsis. Plant Physiol. 2009, 150, 114–124. [Google Scholar] [CrossRef]
- Boynton, A.; Whitfield, J.F.; McManus, J.P. Calmodulin stimulated DNA synthesis by rat liver cells. Biochem. Biophys. Res. Comm. 1980, 95, 745–749. [Google Scholar] [CrossRef]
- Goberdhan, N.J.; Dawson, R.A.; Freedlander, E.E.; MacNeil, E. A calmodulin-like proteins as an extracellular mitogen for the keratinocyte. Brit. J. Dermatol. 1993, 129, 678–688. [Google Scholar] [CrossRef]
- Remgard, P.; Ekstrom, P.A.R.; Wiklund, P.; Edstrom, A. Calmodulin and in vitro in vitro regenerating frog sciatic nerves: Release and extracellular effects. Eur. J. Neurosci. 1995, 7, 1386–1392. [Google Scholar] [CrossRef]
- Ikezaki, H.; Patel, M.; Onyuksel, H.; Akhter, S.R.; Gao, X.P.; Rubenstein, I. Exogenous calmodulin potentiates vasodilation elicited by phospholipid-associated VIP in vivo in vivo. Am. J. Physiol. Reg. Int. Comp. Physiol. 1999, 276, 1359–1365. [Google Scholar]
- O’Day, D.H.; Huber, R.J.; Suarez, A. Extracellular Calmodulin regulates growth and cAMP-mediated chemotaxis in Dictyostelium discoideum. Biochem. Biophys. Res. Commun. 2012, 425, 750–754. [Google Scholar] [CrossRef]
- Gauthier, M.L.; O'Day, D.H. Detection of calmodulin-binding proteins and calmodulin-dependent phosphorylation linked to calmodulin-dependent chemotaxis to folic and cAMP in Dictyostelium. Cell. Signal. 2001, 13, 575–584. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
O'Day, D.H.; Huber, R.J. Matricellular Signal Transduction Involving Calmodulin in the Social Amoebozoan Dictyostelium. Genes 2013, 4, 33-45. https://doi.org/10.3390/genes4010033
O'Day DH, Huber RJ. Matricellular Signal Transduction Involving Calmodulin in the Social Amoebozoan Dictyostelium. Genes. 2013; 4(1):33-45. https://doi.org/10.3390/genes4010033
Chicago/Turabian StyleO'Day, Danton H., and Robert J. Huber. 2013. "Matricellular Signal Transduction Involving Calmodulin in the Social Amoebozoan Dictyostelium" Genes 4, no. 1: 33-45. https://doi.org/10.3390/genes4010033
APA StyleO'Day, D. H., & Huber, R. J. (2013). Matricellular Signal Transduction Involving Calmodulin in the Social Amoebozoan Dictyostelium. Genes, 4(1), 33-45. https://doi.org/10.3390/genes4010033



