A Reverse Transcriptase-Dependent Mechanism Is Essential for Murine Preimplantation Development
Abstract
:1. A Highly Dynamic Genomic Landscape in Preimplantation Embryos
2. The Emergence of Retrotransposition in Developmental Control
3. An Endogenous RT Activity Operates in Spermatozoa and Early Embryos
4. Inhibition of Endogenous RT Arrests Development
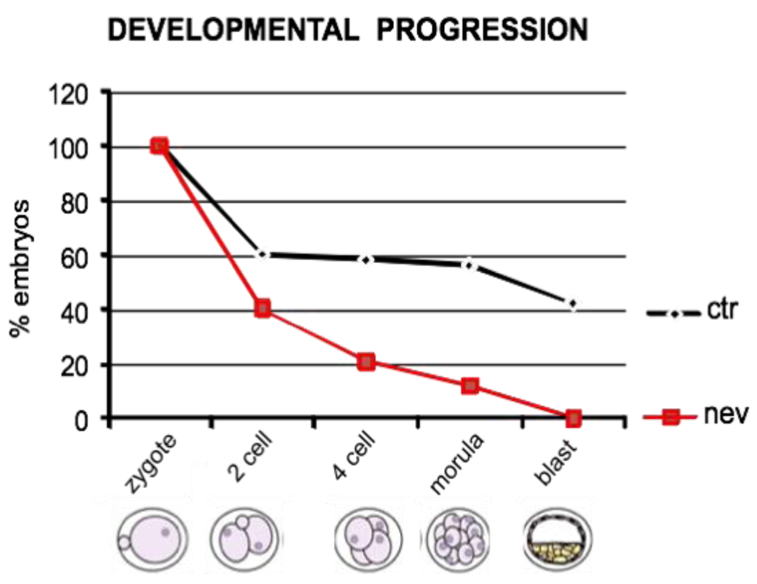
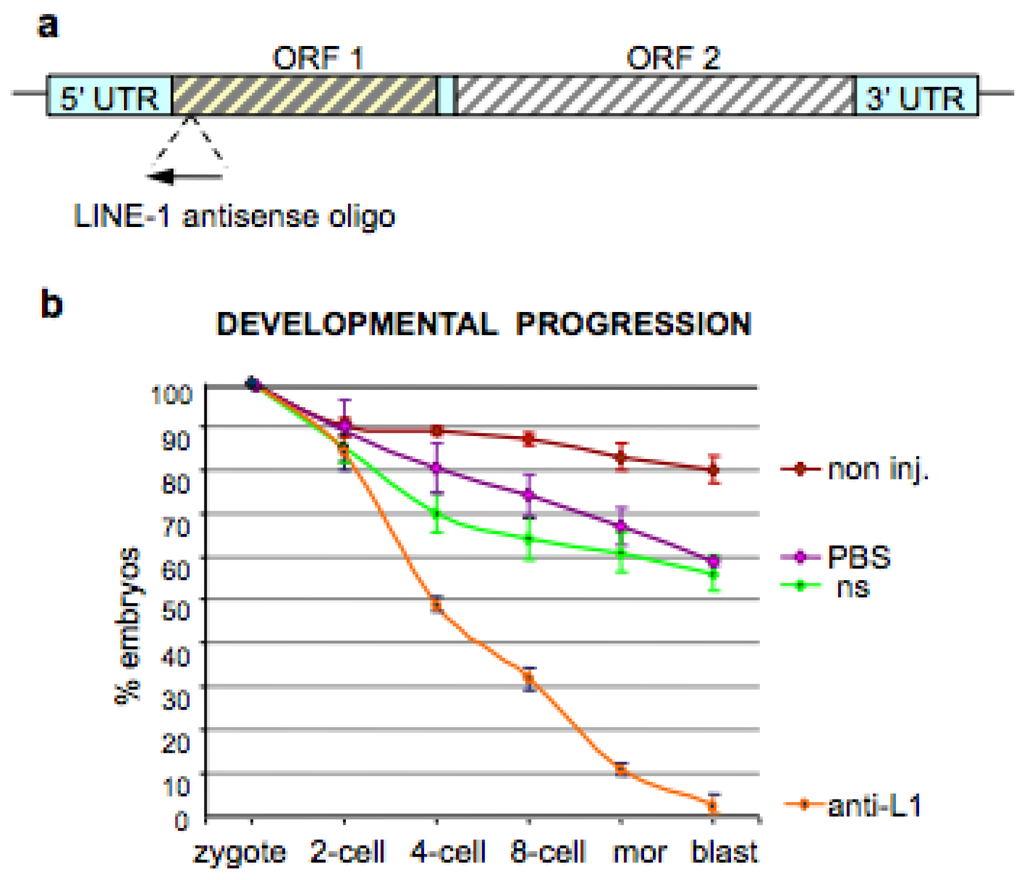
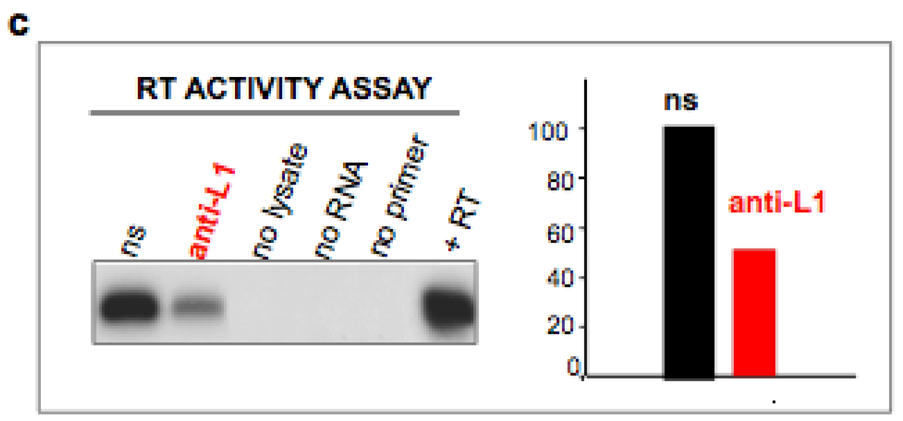
5. The Analogies between Early Embryogenesis and Tumorigenesis
6. A Genome-Wide RT-Dependent Regulatory Mechanism
Acknowledgments
References
- Edwards, R.G.; Beard, H.K. Oocyte polarity and cell determination in early mammalian embryos. Mol. Hum. Reprod. 1997, 3, 863–905. [Google Scholar]
- Fujimori, T.; Kurotaki, Y.; Miyazaki, J.; Nabeshima, Y. Analysis of cell lineage in two- and four-cell mouse embryos. Development 2003, 130, 5113–5122. [Google Scholar]
- Morgan, H.D.; Santos, F.; Green, K.; Dean, W.; Reik, W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005, 14, R47–R58. [Google Scholar]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar]
- Mayer, W.; Smith, A.; Fundele, R.; Haaf, T. Spatial separation of parental genomes in preimplantation mouse embryos. J. Cell Biol. 2000, 148, 629–634. [Google Scholar]
- Haaf, T. The battle of the sexes after fertilization: Behaviour of paternal and maternal chromosomes in the early mammalian embryo. Chromosome Res. 2001, 9, 263–271. [Google Scholar]
- Feil, R. Epigenetic asymmetry in the zigote and mammalian development. Int. J. Dev. Biol. 2009, 53, 191–201. [Google Scholar]
- Ahmed, K.; Dehghani, H.; Rugg-Gunn, P.; Fussner, E.; Rossant, J.; Bazett-Jones, D.P. Global Chromatin Architecture Reflects Pluripotency and Lineage Commitment in the Early Mouse Embryo. PLoS One 2010, 5, e10531. [Google Scholar]
- Tian, W.Q.; Piotrowska, K.; Ciemerych, A.M.; Milenkovic, L.; Scott, M.P.; Davis, R.W.; Zernicka-Goetz, M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 2004, 6, 133–144. [Google Scholar]
- Hamatani, T.; Carter, M.G.; Sharov, A.A.; Ko, M.S.H. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004, 6, 117–131. [Google Scholar]
- Hamatani, T.; Ko, M.S.H.; Yamada, M.; Kuji, N.; Mizusawa, Y.; Shoji, M.; Hada, T.; Asada, H.; Maruyama, T.; Yoshimura, Y. Global gene expression profiling of preimplantation embryos. Human Cell 2006, 19, 98–117. [Google Scholar]
- International Human Genome Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921.
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Nat. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar]
- Goodier, J.L.; Kazazian, H.H. Retrotransposons Revisited: The restraint and rehabilitation of parasites. Cell 2008, 135, 23–35. [Google Scholar]
- Orgel, L.E.; Crick, F.H. Selfish DNA: The ultimate parasite. Nature 1980, 284, 604–607. [Google Scholar]
- Doolittle, W.F.; Sapienza, C. Selfish genes, the phenotype paradigm and genome evolution. Nature 1980, 284, 601–603. [Google Scholar]
- Belancio, V.P.; Hedges, D.J.; Deininger, P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008, 18, 343–358. [Google Scholar]
- Feschotte, C. Transposable elemento and the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar]
- Tomilin, N.V. Regulation of mammalian gene expression by retroelements and non-coding tandem repeats. BioEssays 2008, 30, 338–348. [Google Scholar]
- Peaston, A.E.; Evsikov, A.V.; Graber, J.H.; de Vries, W.N.; Holbrook, A.E.; Solter, D.; Knowles, B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 2004, 7, 597–606. [Google Scholar]
- Evsikov, A.V.; de Vries, W.N.; Peaston, A.E.; Radford, E.E.; Fancher, K.S.; Chen, F.H.; Blake, J.A.; Bult, C.J.; Latham, K.E.; Solter, D.; et al. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 2004, 105, 240–250. [Google Scholar]
- Peaston, A.E.; Knowles, B.B.; Hutchison, K.W. Genome plasticity in the mouse oocyte and early embryo. Biochem. Soc. Trans. 2007, 35, 618–622. [Google Scholar]
- Ohnishi, Y.; Totoki, Y.; Toyoda, A.; Watanabe, T.; Yamamoto, Y.; Tokunaga, K.; Sakaki, Y.; Sasaki, H.; Hohjoh, H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucl. Acid. Res. 2010, 38, 5141–5151. [Google Scholar]
- Poznanski, A.A.; Calarco, P.G. The expression of intracisternal A particle genes in the pre-implantation mouse embryo. Dev. Biol. 1991, 143, 271–281. [Google Scholar]
- Packer, A.I.; Manova, K.; Bacharova, R.F. A discrete LINE-1 transcript in mouse blastocysts. Dev. Biol. 1993, 157, 281–283. [Google Scholar]
- Kigami, D.; Minami, N.; Takayama, H.; Imai, H. MuERV-L, one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod. 2003, 68, 651–654. [Google Scholar]
- van den Hurk, J.A.J.M.; Meij, I.C.; Seleme, M.C.; Kano, H.; Nikopoulos, K.; Hoefsloot, L.H.; Sistermans, E.A.; de Wijs, I.J.; Mukhopadhyay, A.; Plomp, A.S.; et al. L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 2007, 16, 1587–1592. [Google Scholar]
- Garcia-Perez, J.L.; Marchetto, M.C.N.; Muotri, A.R.; Coufal, N.G.; Gage, F.H.; O'Shea, K.S.; Moran, J.V. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007, 16, 1569–1577. [Google Scholar]
- Kano, H.; Godoy, I.; Courtney, C.; Vetter, M.R.; Gerton, G.L.; Ostertag, E.M.; Kazazian, H.H., Jr. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosiaicism. Gene. Dev. 2009, 23, 1303–1312. [Google Scholar]
- Shi, X.; Seluanov, A.; Gorbunova, V. Cell divisions are required for L1 retrotransposition. Mol. Cell Biol. 2007, 27, 1264–1270. [Google Scholar]
- Bourc'his, D.; Bestor, T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar]
- Lane, N.; Dean, W.; Erhardt, S.; Hajkova, P.; Surani, A.; Walter, J.; Reik, W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 2003, 35, 88–93. [Google Scholar]
- Dean, W.; Santos, F.; Reik, W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin. Cell Dev. Biol. 2003, 14, 93–100. [Google Scholar]
- Kim, S.H.; Kang, Y.K.; Koo, D.B.; Kang, M.J.; Moon, S.J.; Lee, K.K.; Han, Y.M. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem. Biophys. Res. Comm. 2004, 324, 58–63. [Google Scholar]
- Svoboda, P.; Stein, P.; Anger, M.; Bernstein, E.; Hannon, G.J.; Schultz, R.M. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 2004, 269, 276–285. [Google Scholar]
- Maksakova, I.A.; Mager, D.L. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J. Virol. 2005, 79, 13865–13874. [Google Scholar]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463, 237–240. [Google Scholar]
- Giordano, R.; Magnano, A.R.; Zaccagnini, G.; Pittoggi, C.; Moscufo, N.; Lorenzini, R.; Spadafora, C. Reverse transcriptase activity in mature spermatozoa of mouse. J. Cell Biol. 2000, 148, 1107–1113. [Google Scholar]
- Pittoggi, C.; Renzi, L.; Zaccagnini, G.; Cimini, D.; Degrassi, F.; Giordano, R.; Magnano, A.R.; Lorenzini, R.; Lavia, P.; Spadafora, C. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J. Cell Sci. 1999, 112, 3537–3548. [Google Scholar]
- Pittoggi, C.; Zaccagnini, G.; Giordano, R.; Magnano, A.R.; Baccetti, B.; Lorenzini, R.; Spadafora, C. Nucleosomal domains of mouse spermatozoa chromatin as potential sites for retroposition and foreign DNA integration. Mol. Reprod. Dev. 2000, 56, 248–251. [Google Scholar]
- Sciamanna, I.; Barberi, L.; Martire, A.; Pittoggi, C.; Beraldi, R.; Giordano, R.; Magnano, A.R.; Hogdson, C.; Spadafora, C. Sperm endogenous reverse transcriptase as mediator of new genetic information. Biochem. Biophys. Res. Commun. 2003, 312, 1039–1046. [Google Scholar]
- Pittoggi, C.; Beraldi, R.; Sciamanna, I.; Barberi, L.; Giordano, R.; Magnano, A.R.; Torosantucci, L.; Pescarmona, E.; Spadafora, C. Generation of biologically active retro-genes upon interaction of mouse spermatozoa with exogenous DNA. Mol. Reprod. Dev. 2006, 73, 1239–1246. [Google Scholar]
- Spadafora, C. A reverse transcriptase-dependent mechanism plays central roles in fundamental biological processes. Syst. Biol. Reprod. Med. 2008, 54, 11–21. [Google Scholar]
- Spadafora, C. Sperm-mediated “reverse” gene transfer: A role of reverse transcriptase in the generation of new genetic information. Hum. Reprod. 2008, 23, 735–740. [Google Scholar]
- Sciamanna, I.; Vitullo, P.; Curatolo, A.; Spadafora, C. Retrotransposons, reverse transcriptase and the genesis of new genetic information. Gene 2009, 448, 180–186. [Google Scholar]
- Pittoggi, C.; Sciamanna, I.; Mattei, E.; Beraldi, R.; Lobascio, A.M.; Mai, A.; Quaglia, M.G.; Lorenzini, R.; Spadafora, C. A role of endogenous reverse transcriptase in murine early embryo development. Mol. Reprod. Dev. 2003, 66, 225–236. [Google Scholar]
- Saunders, C.M.; Larman, M.G.; Parrington, J.; Cox, L.J.; Royse, J.; Blayney, L.M.; Swann, K.; Lai, F.A. PLC zeta: A sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development 2002, 129, 3533–3544. [Google Scholar]
- Ostermeier, G.C.; Miller, D.; Huntriss, J.D.; Diamond, M.P.; Krawetz, S.A. Delivering spermatozoan RNA to the oocyte. Nature 2004, 429, 154. [Google Scholar]
- Barroso, G.; Valdespin, C.; Vega, E.; Kershenovich, R.; Avila, R.; Avendano, C.; Oehninger, S. Developmental sperm contributions: Fertilization and beyond. Fertil. Steril. 2009, 92, 835–848. [Google Scholar]
- Merluzzi, V.I.; Hargrave, K.D.; Labadia, M.; Grozinger, K.; Skoog, M.; Wu, J.C.; Shih, C.K.; Eckner, K.; Hattox, S.; Adams, J.; et al. Inhibition of HIV-1-1 replication by a nonnucleoside reverse transcriptase inhibitor. Science 1990, 250, 1411–1413. [Google Scholar]
- Naas, T.P.; DeBerardinis, R.J.; Moran, J.V.; Ostertag, E.M.; Kingsmore, S.F.; Seldin, M.F.; Hayashizaki, Y.; Martin, S.L.; Kazazian, H.H. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998, 17, 590–597. [Google Scholar]
- Beraldi, R.; Pittoggi, C.; Sciamanna, I.; Mattei, E.; Spadafora, C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 2006, 73, 279–287. [Google Scholar]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W.; et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Gene. Dev. 2008, 22, 908–917. [Google Scholar]
- Virchow, R.L.K. Cellular Pathology, 1859 special ed.; John Churchill: London, UK, 1978; pp. 204–207. [Google Scholar]
- Bailey, P.; Cushing, H. Medulloblastoma cerbelli: A common type of midcerebellar glioma of childhood. Arch. Neurol. Psychiatry 1925, 14, 192–223. [Google Scholar]
- Monk, M.; Holding, C. Human embryonic genes re-expressed in cancer cells. Oncogene 2001, 20, 8085–8091. [Google Scholar]
- Cillo, C.; Faiella, A.; Cantile, M.; Boncinelli, E. Homeobox genes an cancer. Exp. Cell Res. 1999, 248, 1–9. [Google Scholar]
- Owens, B.M.; Hawley, R.G. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells 2002, 20, 364–379. [Google Scholar]
- Samuel, S.; Naoram, H. Homeobox gene expression in cancer: Insights from developmental regulation and deregulation. Eur. J Canc. 2005, 41, 2428–2437. [Google Scholar]
- Ansieau, S.; Bastid, J.; Doreau, A.; Morel, A.P.; Bouchet, B.P.; Thomas, C.; Fauvet, F.; Puisieux, I.; Doglioni, C.; Piccinin, S.; et al. Induction of EMT by twist proteins as a collateral effect of tumor promoting inactivation of premature senescence. Cancer Cell 2008, 14, 79–89. [Google Scholar]
- Boutet, A.; Esteban, M.A.; Maxwell, P.H.; Nieto, M.A. Reactivation of Snail genes in renal fibrosis and carcinomas. Cell Cycle 2007, 66, 638–642. [Google Scholar]
- Schaeffer, E.M.; Marchionni, L.; Huang, Z.; Simons, B.; Blackman, A.; Yu, W.; Parmigiani, G.; Barman, D.M. Androgen induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene 2008, 27, 7180–7191. [Google Scholar]
- Kaiser, S.; Park, Y.K.; Franklin, J.L.; Halberg, L.B.; Yu, M.; Jessen, W.J.; Freudenberg, J.; Chen, X. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007, 8, R131. [Google Scholar]
- Vanneste, E.; Voet, T.; Le Caignec, C.; Ampe, M.; Konings, P.; Melotte, C.; Debrock, S.; Amyere, M.; Vikkula, M.; Schuit, F.; et al. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 2009, 15, 577–583. [Google Scholar]
- Schulz, W.A. L1 Retrotransposons in human cancers. J. Biomed. Biotechnol. 2006, 2006, 1–12. [Google Scholar]
- Ruprecht, K.; Mayer, J.; Sauter, M.; Roemer, K.; Mueller-Lantzsch, N. Endogenous retroviruses and cancer. Cell Mol. Life Sci. 2008, 65, 3366–3382. [Google Scholar]
- Belancio, V.P.; Roy-Engel, A.M.; Deininger, P.L. All “you need to know” bout retroelements in cancer. Semin. Canc. Biol. 2010, 20, 200–210. [Google Scholar]
- Mangiacasale, R.; Pittoggi, C.; Sciamanna, I.; Careddu, A.; Mattei, E.; Lorenzini, R.; Travaglini, L.; Landriscina, M.; Barone, C.; Nervi, C.; et al. Exposure of normal and transformed cells to nevirapine, a Reverse Transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene 2003, 22, 2750–2761. [Google Scholar]
- Sciamanna, I.; Landriscina, M.; Pittoggi, C.; Quirino, M.; Mearelli, C.; Beraldi, R.; Mattei, E.; Serafino, A.; Cassano, A.; Sinibaldi-Vallebona, P.; et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene 2005, 24, 3923–3931. [Google Scholar]
- Landriscina, M.; Fabiano, A.; Altamura, S.; Bagala, C.; Piscazzi, A.; Cassano, A.; Spadafora, C.; Giorgino, F.; Barone, C.; Cignarelli, M. Reverse Transcriptase Inhibitors Down-regulate cell proliferation in vitro and in vivo and restore thyrotropin signaling and iodine uptake in human thyroid anaplastic carcinoma. J. Clin. Endocrinol. Metabol. 2005, 90, 5663–5671. [Google Scholar]
- Oricchio, E.; Sciamanna, I.; Beraldi, R.; Tolstonog, G.V.; Schumann, G.G.; Spadafora, C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene 2007, 26, 4226–4233. [Google Scholar]
- Hagan, C.R.; Rudin, C.M. Mobile genetic element activation and genotoxic cancer therapy potential clinical implications. Am. J. PharmacoGenomics 2002, 2, 25–35. [Google Scholar]
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563–571. [Google Scholar]
- McClintock, B. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 1956, 21, 197–216. [Google Scholar]
- Britten, R.J.; Davidson, E.H. Gene regulation for higher cells: A theory. Science 1969, 165, 349–357. [Google Scholar]
- Britten, R.J.; Davidson, E.H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelties. Q. Rev. Biol. 1971, 46, 111–138. [Google Scholar]
- Temin, H.M. Guest Editorial. J. Natl. Canc. Inst. 1971, 46, III–VI. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sciamanna, I.; Vitullo, P.; Curatolo, A.; Spadafora, C. A Reverse Transcriptase-Dependent Mechanism Is Essential for Murine Preimplantation Development. Genes 2011, 2, 360-373. https://doi.org/10.3390/genes2020360
Sciamanna I, Vitullo P, Curatolo A, Spadafora C. A Reverse Transcriptase-Dependent Mechanism Is Essential for Murine Preimplantation Development. Genes. 2011; 2(2):360-373. https://doi.org/10.3390/genes2020360
Chicago/Turabian StyleSciamanna, Ilaria, Patrizia Vitullo, Angela Curatolo, and Corrado Spadafora. 2011. "A Reverse Transcriptase-Dependent Mechanism Is Essential for Murine Preimplantation Development" Genes 2, no. 2: 360-373. https://doi.org/10.3390/genes2020360
APA StyleSciamanna, I., Vitullo, P., Curatolo, A., & Spadafora, C. (2011). A Reverse Transcriptase-Dependent Mechanism Is Essential for Murine Preimplantation Development. Genes, 2(2), 360-373. https://doi.org/10.3390/genes2020360



