Decoding Non-Coding RNA Regulators in DITRA: From Genomic Insights to Potential Biomarkers and Therapeutic Targets
Abstract
1. Introduction
1.1. Psoriasis Subtypes
1.2. Anosology of Psoriasis
1.3. IL36RN Mutation as the Cause of DITRA
1.4. Molecular Pathway of DITRA
1.5. ncRNAs in DITRA
2. Materials and Methods
2.1. Data Collection and Selection of Non-Coding RNAs
2.2. Non-Coding RNA Evaluation
3. Results
4. Discussion
4.1. Genetic and Epigenetic Context of DITRA and Psoriasis
4.2. Functional Implications of the Newly Discovered Genes in IL36RN Regulation
4.3. Functional Implications of lncRNAs in IL36RN Regulation
4.4. miRNAs in Inflammation and Autoimmunity
4.5. Clinical and Experimental Relevance
4.6. A Molecular Interaction Network Perspective
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DITRA | Deficiency of Interleukin-36 Receptor Antagonist |
| GPP | Generalized Pustular Psoriasis |
| ACH | Hallopeau’s Continuous Acrodermatitis |
| IL36Ra | IL-36 Receptor Antagonist |
| LPP | Localized Pustular Psoriasis |
| MAPKs | Mitogen-activated Protein Kinases |
| NF-kB | Nuclear Factor-Κb |
| PaP | Palmoplantar Psoriasis |
| PP | Pustular Psoriasis |
| PPP | Pustular Psoriasis of Pregnancy |
References
- Raharja, A.; Mahil, S.K.; Barker, J.N. Psoriasis: A brief overview. Clin. Med. 2021, 21, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.A.; Badri, T. Psoriasis. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2025. [Google Scholar]
- Jain, R.; Sangoi, R.; Pascal, S.; Sharma, Y.; Variya Takodara, Y.; Singh, A.A.; Shah, Y.; Prashanth, A.; Agrawal, M.; Wankhede, P. Insights Into the Epidemiological, Clinical, Histopathological, and Dermoscopic Aspects of Chronic Plaque Psoriasis. Cureus 2024, 16, e69912. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.; Mehta, H. Biological treatment for erythrodermic psoriasis. Expert Opin. Biol. Ther. 2022, 22, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Bachelez, H. Pustular Psoriasis: The Dawn of a New Era. Acta Derm.-Venereol. 2020, 100, 5651. [Google Scholar] [CrossRef]
- Shah, M.; Al Aboud, D.M.; Crane, J.S.; Kumar, S. Pustular Psoriasis. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2025. [Google Scholar]
- Marrakchi, S.; Guigue, P.; Renshaw, B.R.; Puel, A.; Pei, X.Y.; Fraitag, S.; Zribi, J.; Bal, E.; Cluzeau, C.; Chrabieh, M.; et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N. Engl. J. Med. 2011, 365, 620–628. [Google Scholar] [CrossRef]
- Liang, Y.; Sarkar, M.K.; Tsoi, L.C.; Gudjonsson, J.E. Psoriasis: A mixed autoimmune and autoinflammatory disease. Curr. Opin. Immunol. 2017, 49, 1–8. [Google Scholar] [CrossRef]
- Krainer, J.; Siebenhandl, S.; Weinhausel, A. Systemic autoinflammatory diseases. J. Autoimmun. 2020, 109, 102421. [Google Scholar] [CrossRef]
- Zormpa, T.; Thireou, T.; Beloukas, A.; Chaniotis, D.; Golfinopoulou, R.; Vlachakis, D.; Eliopoulos, E.; Papageorgiou, L. The Genetic Background of Ankylosing Spondylitis Reveals a Distinct Overlap with Autoimmune Diseases: A Systematic Review. J. Clin. Med. 2025, 14, 3677. [Google Scholar] [CrossRef]
- Furue, K.; Ito, T.; Tsuji, G.; Kadono, T.; Nakahara, T.; Furue, M. Autoimmunity and autoimmune co-morbidities in psoriasis. Immunology 2018, 154, 21–27. [Google Scholar] [CrossRef]
- Okorie, C.L.; Nayudu, K.; Nambudiri, V.E. Cutaneous findings and treatments in deficiency of interleukin-36 receptor antagonist (DITRA): A review of the literature. Exp. Dermatol. 2024, 33, e14934. [Google Scholar] [CrossRef]
- Arakelyan, A.; Nersisyan, L.; Poghosyan, D.; Khondkaryan, L.; Hakobyan, A.; Loffler-Wirth, H.; Melanitou, E.; Binder, H. Autoimmunity and autoinflammation: A systems view on signaling pathway dysregulation profiles. PLoS ONE 2017, 12, e0187572. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.Y.; Huang, Y.W.; Huang, Y.H.; Tsai, T.F. Deficiency of interleukin-36 receptor antagonist (DITRA): An analysis of 58 Chinese patients in a tertiary hospital in Taiwan. Exp. Dermatol. 2023, 32, 1272–1278. [Google Scholar] [CrossRef]
- Bassoy, E.Y.; Towne, J.E.; Gabay, C. Regulation and function of interleukin-36 cytokines. Immunol. Rev. 2018, 281, 169–178. [Google Scholar] [CrossRef]
- Ganesan, R.; Raymond, E.L.; Mennerich, D.; Woska, J.R., Jr.; Caviness, G.; Grimaldi, C.; Ahlberg, J.; Perez, R.; Roberts, S.; Yang, D.; et al. Generation and functional characterization of anti-human and anti-mouse IL-36R antagonist monoclonal antibodies. mAbs 2017, 9, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, C.M.; Marquardt, Y.; Fietkau, K.; Baron, J.M.; Luscher, B. The psoriasis-associated IL-17A induces and cooperates with IL-36 cytokines to control keratinocyte differentiation and function. Sci. Rep. 2017, 7, 15631. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.Y.; Li, L.; Wang, M.J.; Chen, X.M.; Huang, Q.C.; Lu, C.J. An Exploration of the Role of MicroRNAs in Psoriasis: A Systematic Review of the Literature. Medicine 2015, 94, e2030. [Google Scholar] [CrossRef]
- Mathavan, S.; Kue, C.S.; Kumar, S. Identification of potential candidate genes for lip and oral cavity cancer using network analysis. Genom. Inform. 2021, 19, e4. [Google Scholar] [CrossRef]
- Xu, P.; Wu, Q.; Lu, D.; Yu, J.; Rao, Y.; Kou, Z.; Fang, G.; Liu, W.; Han, H. A systematic study of critical miRNAs on cells proliferation and apoptosis by the shortest path. BMC Bioinform. 2020, 21, 396. [Google Scholar] [CrossRef]
- Yuan, Z.C.; Xu, W.D.; Liu, X.Y.; Liu, X.Y.; Huang, A.F.; Su, L.C. Biology of IL-36 Signaling and Its Role in Systemic Inflammatory Diseases. Front. Immunol. 2019, 10, 2532. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Killcoyne, S.; Carter, G.W.; Smith, J.; Boyle, J. Cytoscape: A community-based framework for network modeling. In Protein Networks and Pathway Analysis; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 563, pp. 219–239. [Google Scholar] [CrossRef]
- Kang, J.; Tang, Q.; He, J.; Li, L.; Yang, N.; Yu, S.; Wang, M.; Zhang, Y.; Lin, J.; Cui, T.; et al. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res. 2022, 50, D326–D332. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Chen, Y.; Zhang, M.; Xu, D.Q.; Liu, Y.; Liu, T.; Liu, S.X.; Wang, P. Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis. PeerJ 2018, 6, e5180. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, R.; Dai, T.; Guo, Y.; Tian, Z.; Zhu, Y.; Chen, J.; Yu, Y. Identification of hub genes and key pathways in arsenic-treated rice (Oryza sativa L.) based on 9 topological analysis methods of CytoHubba. Environ. Health Prev. Med. 2024, 29, 41. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef]
- Zhang, S.; Amahong, K.; Zhang, Y.; Hu, X.; Huang, S.; Lu, M.; Zeng, Z.; Li, Z.; Zhang, B.; Qiu, Y.; et al. RNAenrich: A web server for non-coding RNA enrichment. Bioinformatics 2023, 39, btad421. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef] [PubMed]
- Onoufriadis, A.; Simpson, M.A.; Pink, A.E.; Di Meglio, P.; Smith, C.H.; Pullabhatla, V.; Knight, J.; Spain, S.L.; Nestle, F.O.; Burden, A.D.; et al. Mutations in IL36RN/IL1F5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. Am. J. Hum. Genet. 2011, 89, 432–437. [Google Scholar] [CrossRef]
- Sugiura, K.; Takemoto, A.; Yamaguchi, M.; Takahashi, H.; Shoda, Y.; Mitsuma, T.; Tsuda, K.; Nishida, E.; Togawa, Y.; Nakajima, K.; et al. The majority of generalized pustular psoriasis without psoriasis vulgaris is caused by deficiency of interleukin-36 receptor antagonist. J. Investig. Dermatol. 2013, 133, 2514–2521. [Google Scholar] [CrossRef]
- Hussain, S.; Berki, D.M.; Choon, S.E.; Burden, A.D.; Allen, M.H.; Arostegui, J.I.; Chaves, A.; Duckworth, M.; Irvine, A.D.; Mockenhaupt, M.; et al. IL36RN mutations define a severe autoinflammatory phenotype of generalized pustular psoriasis. J. Allergy Clin. Immunol. 2015, 135, 1067–1070.e9. [Google Scholar] [CrossRef] [PubMed]
- Tauber, M.; Bal, E.; Pei, X.Y.; Madrange, M.; Khelil, A.; Sahel, H.; Zenati, A.; Makrelouf, M.; Boubridaa, K.; Chiali, A.; et al. IL36RN Mutations Affect Protein Expression and Function: A Basis for Genotype-Phenotype Correlation in Pustular Diseases. J. Investig. Dermatol. 2016, 136, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Korber, A.; Mossner, R.; Renner, R.; Sticht, H.; Wilsmann-Theis, D.; Schulz, P.; Sticherling, M.; Traupe, H.; Huffmeier, U. Mutations in IL36RN in patients with generalized pustular psoriasis. J. Investig. Dermatol. 2013, 133, 2634–2637. [Google Scholar] [CrossRef]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [PubMed]
- Espe, S. Malacards: The Human Disease Database. J. Med Libr. Assoc. 2018, 106, 140–141. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Kielbowski, K.; Jedrasiak, A.; Bakinowska, E.; Pawlik, A. The Role of Long Non-Coding RNA in the Pathogenesis of Psoriasis. Non-Coding RNA 2025, 11, 7. [Google Scholar] [CrossRef]
- Song, J.K.; Yin, S.Y.; Li, W.; Li, X.D.; Luo, Y.; Luo, Y.; Xing, M.; Li, B.; Kuai, L. An update on the role of long non-coding RNAs in psoriasis. Chin. Med. J. 2020, 134, 379–389. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, J.; Chen, X.; Xia, H.; Yang, L. E3 ubiquitin ligase Trim33 ubiquitylates Annexin A2 to promote NF-kappaB induced skin inflammation in psoriasis. J. Dermatol. Sci. 2022, 107, 160–168. [Google Scholar] [CrossRef]
- Baran, A.; Swiderska, M.; Bacharewicz-Szczerbicka, J.; Mysliwiec, H.; Flisiak, I. Serum Fatty Acid-Binding Protein 4 is Increased in Patients with Psoriasis. Lipids 2017, 52, 51–60. [Google Scholar] [CrossRef]
- Gupta, R.; Debbaneh, M.G.; Liao, W. Genetic Epidemiology of Psoriasis. Curr. Dermatol. Rep. 2014, 3, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Zima, K.; Purzycka-Bohdan, D.; Szczerkowska-Dobosz, A.; Gabig-Ciminska, M. Keratinocyte-Mediated Antigen Presentation in Psoriasis: Preliminary Insights from In Vitro Studies. Int. J. Mol. Sci. 2024, 25, 13387. [Google Scholar] [CrossRef]
- Goodman, L.D.; Cope, H.; Nil, Z.; Ravenscroft, T.A.; Charng, W.L.; Lu, S.; Tien, A.C.; Pfundt, R.; Koolen, D.A.; Haaxma, C.A.; et al. TNPO2 variants associate with human developmental delays, neurologic deficits, and dysmorphic features and alter TNPO2 activity in Drosophila. Am. J. Hum. Genet. 2021, 108, 1669–1691. [Google Scholar] [CrossRef] [PubMed]
- Dopytalska, K.; Ciechanowicz, P.; Wiszniewski, K.; Szymanska, E.; Walecka, I. The Role of Epigenetic Factors in Psoriasis. Int. J. Mol. Sci. 2021, 22, 9294. [Google Scholar] [CrossRef]
- Shefler, A.; Patrick, M.T.; Wasikowski, R.; Chen, J.; Sarkar, M.K.; Gudjonsson, J.E.; Tsoi, L.C. Skin-Expressing lncRNAs in Inflammatory Responses. Front. Genet. 2022, 13, 835740. [Google Scholar] [CrossRef]
- Ghahramani Almanghadim, H.; Karimi, B.; Valizadeh, S.; Ghaedi, K. Biological functions and affected signaling pathways by Long Non-Coding RNAs in the immune system. Non-Coding RNA Res. 2025, 10, 70–90. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Ma, R.; Jiang, X.; Tang, X.; Gong, Y.; Yu, Z.; Shi, Y. Implications of LncRNAs and CircRNAs in psoriasis: A review. RNA Biol. 2023, 20, 334–347. [Google Scholar] [CrossRef]
- Yang, J.; Wang, S.; Liu, L.; Wang, J.; Shao, Y. Long non-coding RNA NEAT1 and its targets (microRNA-21 and microRNA-125a) in rheumatoid arthritis: Altered expression and potential to monitor disease activity and treatment outcome. J. Clin. Lab. Anal. 2021, 35, e24076. [Google Scholar] [CrossRef]
- Wang, D.; Cheng, S.; Zou, G.; Ding, X. Paeoniflorin inhibits proliferation and migration of psoriatic keratinocytes via the lncRNA NEAT1/miR-3194-5p/Galectin-7 axis. Anti-Cancer Drugs 2022, 33, e423–e433. [Google Scholar] [CrossRef]
- Alhelf, M.; Rashed, L.; Doss, R.W.; Mohamed, S.M.; Abd Elazeem, N.A. Long noncoding RNA (taurine upregulated gene 1) and micro RNA-377: Emerging players in the development of metabolic syndrome among psoriasis patients. Beni-Suef Univ. J. Basic Appl. Sci. 2023, 12, 68. [Google Scholar] [CrossRef]
- Zhang, W.; Yi, X.; An, Y.; Guo, S.; Li, S.; Song, P.; Chang, Y.; Zhang, S.; Gao, T.; Wang, G.; et al. MicroRNA-17-92 cluster promotes the proliferation and the chemokine production of keratinocytes: Implication for the pathogenesis of psoriasis. Cell Death Dis. 2018, 9, 567. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, D.; Zhan, M.; Ding, H.; Zhao, H. MicroRNA-146a and microRNA-146b deficiency correlates with exacerbated disease activity, and their longitude increment relates to etanercept response in psoriasis patients. J. Clin. Lab. Anal. 2022, 36, e24198. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Liu, C.; Chen, X.; Li, P.; Qiu, W.; Guo, K. Integrative analysis of gene and microRNA expression profiles reveals candidate biomarkers and regulatory networks in psoriasis. Medicine 2024, 103, e39002. [Google Scholar] [CrossRef]
- Alatas, E.T.; Kara, M.; Dogan, G.; Akin Belli, A. Blood microRNA expressions in patients with mild to moderate psoriasis and the relationship between microRNAs and psoriasis activity. An. Bras. Dermatol. 2020, 95, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Laha, S.; Das, S.; Banerjee, U.; Ganguly, T.; Senapati, S.; Chatterjee, G.; Chatterjee, R. Genome-wide RNA-seq, DNA methylation and small RNA-seq analysis unraveled complex gene regulatory networks in psoriasis pathogenesis. Gene 2025, 933, 148903. [Google Scholar] [CrossRef]
- Delic, D.; Wolk, K.; Schmid, R.; Gabrielyan, O.; Christou, D.; Rieber, K.; Rolser, M.; Jakob, I.; Wiech, F.; Griesser, M.; et al. Integrated microRNA/mRNA expression profiling of the skin of psoriasis patients. J. Dermatol. Sci. 2020, 97, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Torri, A.; Carpi, D.; Bulgheroni, E.; Crosti, M.C.; Moro, M.; Gruarin, P.; Rossi, R.L.; Rossetti, G.; Di Vizio, D.; Hoxha, M.; et al. Extracellular MicroRNA Signature of Human Helper T Cell Subsets in Health and Autoimmunity. J. Biol. Chem. 2017, 292, 2903–2915. [Google Scholar] [CrossRef]
- Jiang, M.; Ma, W.; Gao, Y.; Jia, K.; Zhang, Y.; Liu, H.; Sun, Q. IL-22-induced miR-122-5p promotes keratinocyte proliferation by targeting Sprouty2. Exp. Dermatol. 2017, 26, 368–374. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, C.; Zeng, B.; Tang, X.; Zhang, Y.; Xiang, L.; Mi, L.; Pan, Y.; Wang, H.; Yang, Z. miR124-3p/FGFR2 axis inhibits human keratinocyte proliferation and migration and improve the inflammatory microenvironment in psoriasis. Mol. Immunol. 2020, 122, 89–98. [Google Scholar] [CrossRef]
- Pelosi, A.; Lunardi, C.; Fiore, P.F.; Tinazzi, E.; Patuzzo, G.; Argentino, G.; Moretta, F.; Puccetti, A.; Dolcino, M. MicroRNA Expression Profiling in Psoriatic Arthritis. BioMed Res. Int. 2018, 2018, 7305380. [Google Scholar] [CrossRef]
- Pierouli, K.; Papageorgiou, L.; Chrousos, G.P.; Eliopoulos, E.; Vlachakis, D. Role of lncRNAs related to NRs in the regulation of gene expression. Int. J. Epigenet. 2025, 5, 3. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]

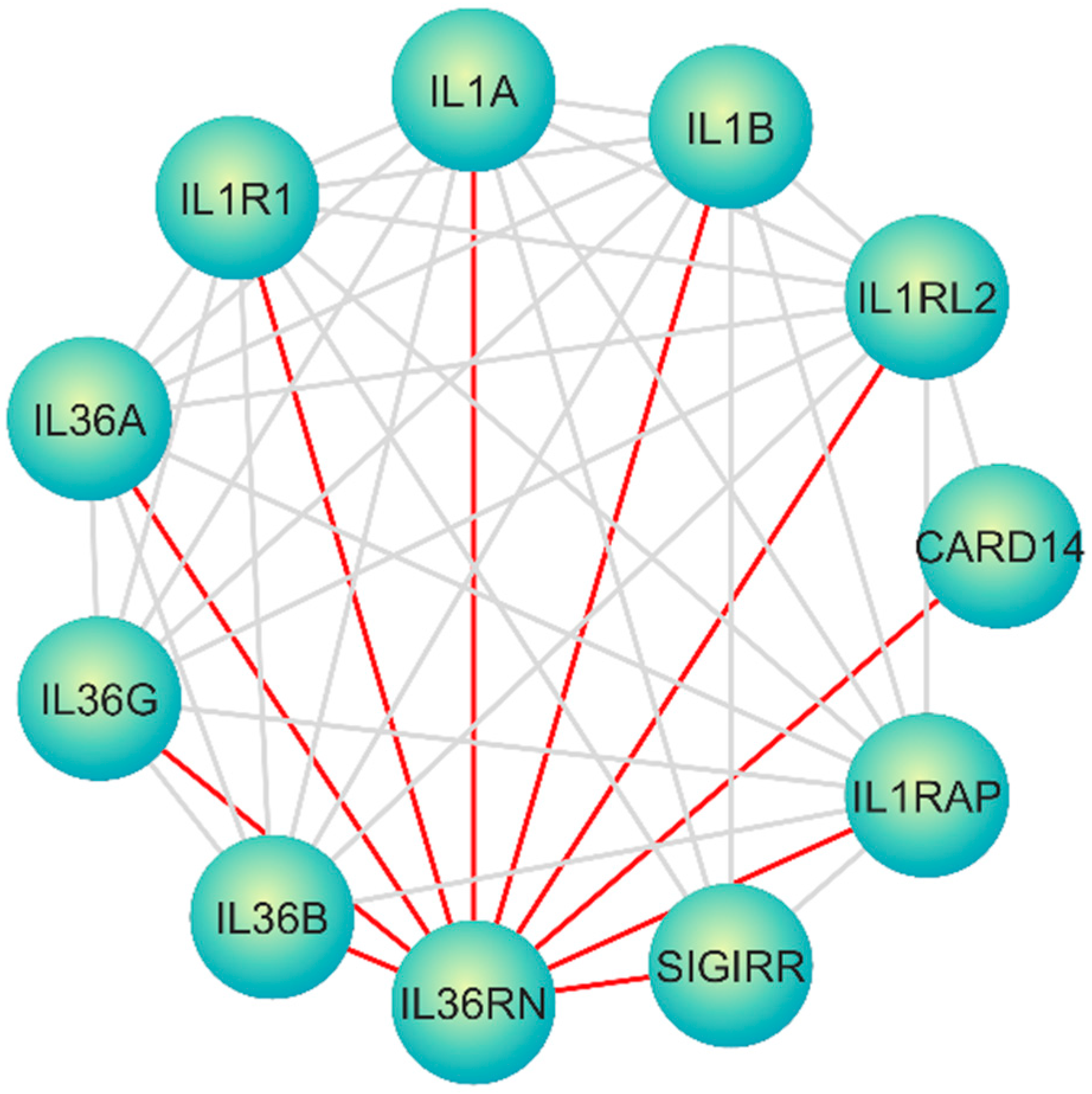
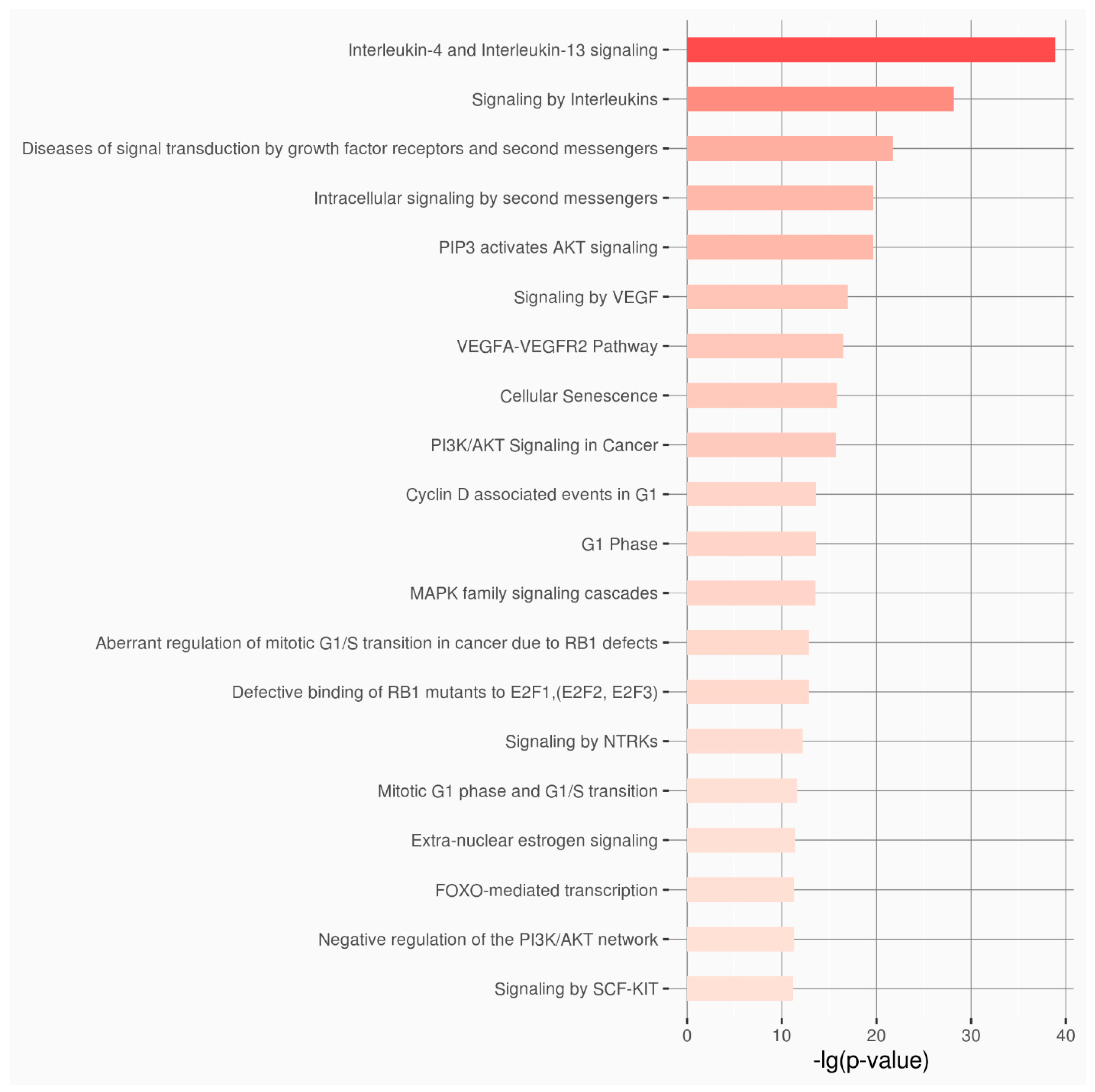
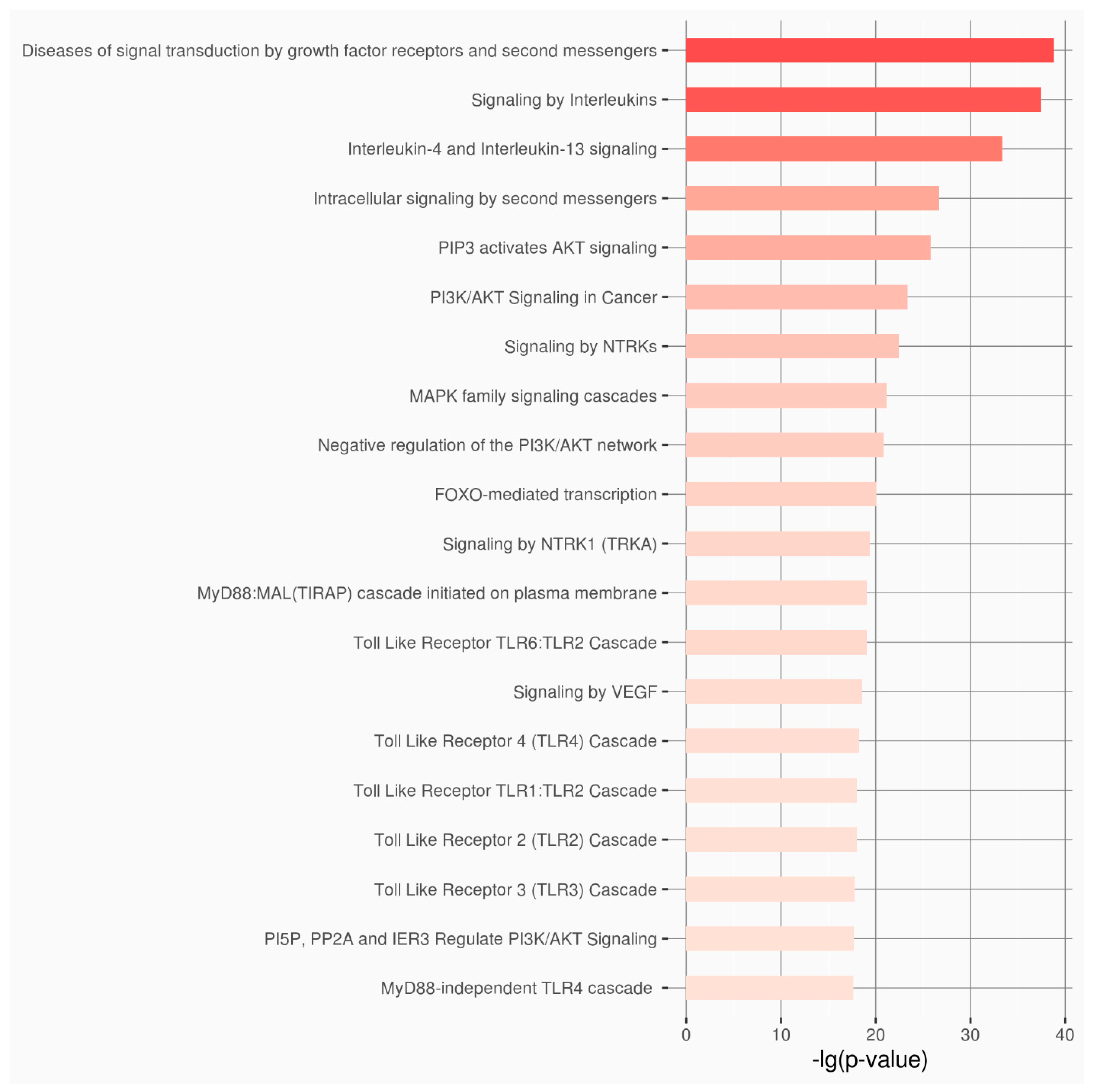
| Category of Functional Genomic Regions | Total |
|---|---|
| miRNA | 333 |
| Genes | 120 |
| lncRNA | 36 |
| Pseudogenes | 9 |
| snRNA | 6 |
| rRNA | 3 |
| eRNA | 2 |
| Unknown Regions | 2 |
| snoRNA | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spanou, S.; Andreou, A.; Gioti, K.; Chaniotis, D.; Beloukas, A.; Papageorgiou, L.; Thireou, T. Decoding Non-Coding RNA Regulators in DITRA: From Genomic Insights to Potential Biomarkers and Therapeutic Targets. Genes 2025, 16, 753. https://doi.org/10.3390/genes16070753
Spanou S, Andreou A, Gioti K, Chaniotis D, Beloukas A, Papageorgiou L, Thireou T. Decoding Non-Coding RNA Regulators in DITRA: From Genomic Insights to Potential Biomarkers and Therapeutic Targets. Genes. 2025; 16(7):753. https://doi.org/10.3390/genes16070753
Chicago/Turabian StyleSpanou, Sofia, Athena Andreou, Katerina Gioti, Dimitrios Chaniotis, Apostolos Beloukas, Louis Papageorgiou, and Trias Thireou. 2025. "Decoding Non-Coding RNA Regulators in DITRA: From Genomic Insights to Potential Biomarkers and Therapeutic Targets" Genes 16, no. 7: 753. https://doi.org/10.3390/genes16070753
APA StyleSpanou, S., Andreou, A., Gioti, K., Chaniotis, D., Beloukas, A., Papageorgiou, L., & Thireou, T. (2025). Decoding Non-Coding RNA Regulators in DITRA: From Genomic Insights to Potential Biomarkers and Therapeutic Targets. Genes, 16(7), 753. https://doi.org/10.3390/genes16070753







