Methods for Extracellular Vesicle Isolation: Relevance for Encapsulated miRNAs in Disease Diagnosis and Treatment
Abstract
1. Introduction
2. EVs: Biogenesis, Function, and Clinical Potential
2.1. Biogenesis of EVs
2.2. EV Function
2.3. EVs Clinical Potential
3. Methods to Isolate EVs
3.1. UC
3.1.1. dUC
3.1.2. DGUC
3.2. Precipitation
3.3. Immunoaffinity
3.4. FACSCanto and EV Sorting
3.5. UF
3.6. SEC
3.7. Microfluidics
3.8. EV Isolation Methods Overview
3.9. EV Quantification and Characterization
4. Biogenesis and Function of microRNAs
4.1. microRNA Biogenesis
4.2. microRNA Function
4.3. microRNAs in Therapy
4.4. microRNAs in Diagnosis
5. Methods to Assess miRNA Levels
5.1. Quantitative Real-Time PCR (qRT-PCR)
5.2. Microarray Analysis
5.3. Next-Generation Sequencing (NGS)
5.4. In Situ Hybridization
5.5. Northern Blotting
5.6. Biosensors
5.7. Digital Droplet PCR (ddPCR)
5.8. NanoString
5.9. Overview of Methods to Quantify miRNA Levels
6. microRNAs Encapsulated in Extracellular Vesicle in Diagnosis and Treatment
| Biomarker | Associated Disease | Isolation and Detection Methodology | Reference |
|---|---|---|---|
| miR-21, miR-126, miR-146a | COVID-19 | UC for EV isolation; qRT-PCR for miRNA detection | [266] |
| miR-21, miR-155 | Lung Cancer | UC for EV isolation; qRT-PCR for miRNA detection | [29] |
| miR-122, miR-192 | Hepatocellular Carcinoma | UC for EV isolation; NGS for miRNA detection | [267] |
| miR-29a, miR-181b | Alzheimer’s Disease | UC for EV isolation; Surface-Enhanced Raman Scattering (SERS) for miRNA detection | [268] |
| miR-21, miR-141 | Prostate Cancer | UC for EV isolation; qRT-PCR for miRNA detection | [269] |
| miR-21, miR-1246 | Esophageal Squamous Cell Carcinoma | Glycosylated EV capture strategy; qRT-PCR for miRNA detection | [270] |
| miR-155, miR-210 | Diffuse Large B-Cell Lymphoma | UC for EV isolation; qRT-PCR for miRNA detection | [271] |
| miR-21, miR-29a | Colorectal Cancer | UC for EV isolation; qRT-PCR for miRNA detection | [272] |
| miR-1246, miR-4644 | Pancreatic Cancer | UC for EV isolation; qRT-PCR for miRNA detection | [273] |
| miR-21, miR-221 | Glioblastoma | UC for EV isolation; qRT-PCR for miRNA detection | [274] |
Clinical Status
7. Discussion
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| EV | Extracellular vesicle |
| miRNA | microRNA |
| MVBs | Multivesicular bodies |
| ESCRT | Endosomal Sorting Complexes Required for Transport |
| MSC | Mesenchymal Stem Cell |
| UC | Ultracentrifugation |
| dUC | Differential Ultracentrifugation |
| DGUC | Density Gradient Ultracentrifugation |
| PEG | Polyethylene Glycol |
| TEIR | Total Exosome Isolation Reagent |
| FSC | Forward Scattered |
| SSC | Side Scattered |
| sEV | Small extracellular vesicle |
| MWCO | Molecular Weight Cutoff |
| UF | Ultrafiltration |
| TFF | Tangential Flow Filtration |
| SEC | Size exclusion chromatography |
| RInSE | Rapid Inertial Solution Exchange |
| PEEK | Polyetheretherketone |
| TBS | Tris-Buffered Saline |
| IgG | Immunoglobulin G |
| pre-miRNA | miRNA precursor |
| DGCR8 | DiGeorge Syndrome Critical Region 8 |
| RISC | RNA-induced silencing complex |
| mRNA | messenger RNA |
| Ago2 | Argonaute 2 |
| RBP | RNA-binding protein |
| lncRNAs | long non-coding RNAs |
| UTR | Untranslated Region |
| ASO | Antisense Oligonucleotide |
| HCV | Hepatitis C Virus |
| qPCR | quantitative Polymerase Chain Reaction |
| qRT-PCR | Quantitative Real-Time PCR |
| RT | reverse transcription |
| cDNA | complementary DNA |
| NGS | Next-Generation Sequencing |
| ISH | In Situ Hybridization |
| smRNA | Single-molecule RNA |
| NB | Northern Blot |
| SERS | Surface-enhanced Raman scattering |
| EM | Electromagnetic Mechanism |
| CM | Chemical-enhancement Mechanism |
| PI-SPR | Phase Imaging Surface Plasmon Resonance |
| ddPCR | Digital Droplet PCR |
| MS | Mass Spectrometry |
| LC | Liquid Chromatography |
| DLS | Dynamic Light Scattering |
| NTA | Nanoparticle Tracking Analysis |
| AFM | Atomic Force Microscopy |
| TIRF-M | Total Internal Reflection Microscopy |
| SEM | Scanning Electron Microscopy |
| TEM | Transmission Electron Microscopy |
| cryo-TEM | Cryo-Transmission Electron Microscopy |
| TRPS | Tunable Resistive Pulse Sensing |
| IRIS | Interference Reflectance Imaging Sensor |
| SMLM | Single-Molecule Localization Microscopy |
| STED | Stimulated Emission Depletion |
| RS | Raman Spectroscopy |
| SERS | Surface-enhanced Raman scattering |
| TERS | Tip-Enhanced Raman Scattering |
| FT-IR | Fourier-Transform Infrared |
| FCS | Fluorescence Correlation Spectroscopy |
| micro-CT | Micro-Computed Tomography |
| ELISA | Enzyme-Linked Immunosorbent Assay |
References
- Das, S.; Lyon, C.J.; Hu, T. A Panorama of Extracellular Vesicle Applications: From Biomarker Detection to Therapeutics. ACS Nano 2024, 18, 9784–9797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, L.; Zhang, X.; Yang, Z.; Wang, B.; Gong, H.; Zhang, K.; Lin, Y.; Sun, M. Extracellular Vesicles: Biological Mechanisms and Emerging Therapeutic Opportunities in Neurodegenerative Diseases. Transl. Neurodegener. 2024, 13, 60. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Su, Y.; Sharma, M.; Singh, S.; Kim, S.; Peavey, J.J.; Suerken, C.K.; Lockhart, S.N.; Whitlow, C.T.; Craft, S.; et al. MicroRNA expression in extracellular vesicles as a novel blood-based biomarker for Alzheimer’s disease. Alzheimer Dement. 2023, 19, 4952–4966. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.H.; Kim, S.Y.; Rudge, C.; Chrzanowski, W. Made by cells for cells—Extracellular vesicles as next-generation mainstream medicines. J. Cell Sci. 2022, 135, jcs259166. [Google Scholar] [CrossRef]
- EL Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Mustajab, T.; Kwamboka, M.S.; Choi, D.A.; Kang, D.W.; Kim, J.; Han, K.R.; Han, Y.; Lee, S.; Song, D.; Chwae, Y.J. Update on Extracellular Vesicle-Based Vaccines and Therapeutics to Combat COVID-19. Int. J. Mol. Sci. 2022, 23, 11247. [Google Scholar] [CrossRef]
- Bajo-Santos, C.; Brokāne, A.; Zayakin, P.; Endzeliņš, E.; Soboļevska, K.; Belovs, A.; Jansons, J.; Sperga, M.; Llorente, A.; Radoviča-Spalviņa, I.; et al. Plasma and urinary extracellular vesicles as a source of RNA biomarkers for prostate cancer in liquid biopsies. Front. Mol. Biosci. 2023, 10, 980433. [Google Scholar] [CrossRef]
- Makarova, J.; Turchinovich, A.; Shkurnikov, M.; Tonevitsky, A. Extracellular miRNAs and Cell-Cell Communication: Problems and Prospects. Trends Biochem. Sci. 2021, 46, 640–651. [Google Scholar] [CrossRef]
- Zeng, E.Z.; Chen, I.; Chen, X.; Yuan, X. Exosomal MicroRNAs as Novel Cell-Free Therapeutics in Tissue Engineering and Regenerative Medicine. Biomedicines 2022, 10, 2485. [Google Scholar] [CrossRef]
- Xu, K.; Liu, Q.; Wu, K.; Liu, L.; Zhao, M.; Yang, H.; Wang, X.; Wang, W. Extracellular vesicles as potential biomarkers and therapeutic approaches in autoimmune diseases. J. Transl. Med. 2020, 18, 432. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Doyle, L.M.; Wang, M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, E.; Boing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef]
- Li, S.R.; Man, Q.W.; Gao, X.; Lin, H.; Wang, J.; Su, F.C.; Wang, H.Q.; Bu, L.L.; Liu, B.; Chen, G. Tissue-derived extracellular vesicles in cancers and non-cancer diseases: Present and future. J. Extracell. Vesicles 2021, 10, e12175. [Google Scholar] [CrossRef]
- Bång-Rudenstam, A.; Cerezo-Magaña, M.; Belting, M. Pro-metastatic functions of lipoproteins and extracellular vesicles in the acidic tumor microenvironment. Cancer Metastasis Rev. 2019, 38, 79–92. [Google Scholar] [CrossRef]
- Gu, J.; Chu, X.; Huo, Y.; Liu, C.; Chen, Q.; Hu, S.; Pei, Y.; Ding, P.; Pang, S.; Wang, M. Gastric cancer-derived exosomes facilitate pulmonary metastasis by activating ERK-mediated immunosuppressive macrophage polarization. J. Cell. Biochem. 2023, 124, 557–572. [Google Scholar] [CrossRef]
- Wilczak, M.; Surman, M.; Przybyło, M. Melanoma-derived extracellular vesicles transfer proangiogenic factors. Oncol. Res. 2025, 33, 245–262. [Google Scholar] [CrossRef]
- Chaput, N.; Thery, C. Exosomes: Immune properties and potential clinical implementations. Semin. Immunopathol. 2011, 33, 419–440. [Google Scholar] [CrossRef]
- Fu, P.; Yin, S.; Cheng, H.; Xu, W.; Jiang, J. Engineered Exosomes for Drug Delivery in Cancer Therapy: A Promising Approach and Application. Curr. Drug Deliv. 2024, 21, 817–827. [Google Scholar] [CrossRef]
- Creeden, J.F.; Sevier, J.; Zhang, J.-T.; Lapitsky, Y.; Brunicardi, F.C.; Jin, G.; Nemunaitis, J.; Liu, J.-Y.; Kalinoski, A.; Rao, D.; et al. Smart exosomes enhance PDAC targeted therapy. J. Control. Release Off. J. Control. Release Soc. 2024, 368, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Gong, H.; Luo, S.; Cui, Y. The Role of Exosomes and Their Applications in Cancer. Int. J. Mol. Sci. 2021, 22, 12204. [Google Scholar] [CrossRef] [PubMed]
- Zanirati, G.; dos Santos, P.G.; Alcará, A.M.; Bruzzo, F.; Ghilardi, I.M.; Wietholter, V.; Xavier, F.A.C.; Gonçalves, J.I.B.; Marinowic, D.; Shetty, A.K.; et al. Extracellular Vesicles: The Next Generation of Biomarkers and Treatment for Central Nervous System Diseases. Int. J. Mol. Sci. 2024, 25, 7371. [Google Scholar] [CrossRef] [PubMed]
- Rolle, K.; Piwecka, M.; Belter, A.; Wawrzyniak, D.; Jeleniewicz, J.; Barciszewska, M.Z.; Barciszewski, J. The Sequence and Structure Determine the Function of Mature Human miRNAs. PLoS ONE 2016, 11, e0151246. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Slezak-Prochazka, I.; Durmus, S.; Kroesen, B.J.; van den Berg, A. MicroRNAs, macrocontrol: Regulation of miRNA processing. RNA 2010, 16, 1087–1095. [Google Scholar] [CrossRef]
- Mustafa, R.; Mens, M.M.J.; van Hilten, A.; Huang, J.; Roshchupkin, G.; Huan, T.; Broer, L.; van Meurs, J.B.J.; Elliott, P.; Levy, D.; et al. A comprehensive study of genetic regulation and disease associations of plasma circulatory microRNAs using population-level data. Genome Biol. 2024, 25, 276. [Google Scholar] [CrossRef]
- Tiwari, P.K.; Shanmugam, P.; Karn, V.; Gupta, S.; Mishra, R.; Rustagi, S.; Chouhan, M.; Verma, D.; Jha, N.K.; Kumar, S. Extracellular Vesicular miRNA in Pancreatic Cancer: From Lab to Therapy. Cancers 2024, 16, 2179. [Google Scholar] [CrossRef]
- Lv, J.; Xiong, X. Extracellular Vesicle microRNA: A Promising Biomarker and Therapeutic Target for Respiratory Diseases. Int. J. Mol. Sci. 2024, 25, 9147. [Google Scholar] [CrossRef]
- Asleh, K.; Dery, V.; Taylor, C.; Davey, M.; Djeungoue-Petga, M.A.; Ouellette, R.J. Extracellular vesicle-based liquid biopsy biomarkers and their application in precision immuno-oncology. Biomark. Res. 2023, 11, 99. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, S.; Yang, D.; Xu, W.; Qian, H. Extracellular vesicles: Emerging roles, biomarkers and therapeutic strategies in fibrotic diseases. J. Nanobiotechnol. 2023, 21, 164. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Anderson, H.C. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J. Cell Biol. 1969, 41, 59–72. [Google Scholar] [CrossRef]
- De Broe, M.; Wieme, R.; Roels, F. Letter: Membrane fragments with koinozymic properties released from villous adenoma of the rectum. Lancet 1975, 306, 1214–1215. [Google Scholar] [CrossRef]
- Benz, E.W., Jr.; Moses, H.L. Small, virus-like particles detected in bovine sera by electron microscopy. J. Natl. Cancer Inst. 1974, 52, 1931–1934. [Google Scholar] [CrossRef]
- Dalton, A.J. Microvesicles and vesicles of multivesicular bodies versus ‘‘virus-like’’ particles. J. Natl. Cancer Inst. 1975, 54, 1137–1148. [Google Scholar] [CrossRef]
- Stegmayr, B.; Ronquist, G. Promotive effect on human sperm progressive motility by prostasomes. Urol. Res. 1982, 10, 253–257. [Google Scholar] [CrossRef]
- Taylor, D.D.; Homesley, H.D.; Doellgast, G.J. Binding of specific peroxidase-labeled antibody to placental-type phosphatase on tumor-derived membrane fragments. Cancer Res. 1980, 40, 4064–4069. [Google Scholar]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P.-K. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Borges, F.; Reis, L.; Schor, N. Extracellular vesicles: Structure, function, and potential clinical uses in renal diseases. Braz. J. Med. Biol. Res. 2013, 46, 824–830. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef]
- Verderio, C.; Gabrielli, M.; Giussani, P. Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J. Lipid Res. 2018, 59, 1325–1340. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Reinisch, K.; Ferro-Novick, S. Coats, Tethers, Rabs, and SNAREs Work Together to Mediate the Intracellular Destination of a Transport Vesicle. Dev. Cell 2007, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aloi, N.; Drago, G.; Ruggieri, S.; Cibella, F.; Colombo, P.; Longo, V. Extracellular Vesicles and Immunity: At the Crossroads of Cell Communication. Int. J. Mol. Sci. 2024, 25, 1205. [Google Scholar] [CrossRef]
- Essien, S.A.; Ahuja, I.; Eisenhoffer, G.T. Apoptotic extracellular vesicles carrying Mif regulate macrophage recruitment and compensatory proliferation in neighboring epithelial stem cells during tissue maintenance. PLoS Biol. 2024, 22, e3002194. [Google Scholar] [CrossRef]
- Zheng, T.; Pu, J.; Chen, Y.; Mao, Y.; Guo, Z.; Pan, H.; Zhang, L.; Zhang, H.; Sun, B.; Zhang, B. Plasma Exosomes Spread and Cluster Around β-Amyloid Plaques in an Animal Model of Alzheimer’s Disease. Front. Aging Neurosci. 2017, 9, 1227. [Google Scholar] [CrossRef]
- Zhang, X.; Che, X.; Zhang, S.; Wang, R.; Li, M.; Jin, Y.; Wang, T.; Song, Y. Mesenchymal stem cell-derived extracellular vesicles for human diseases. Extracell. Vesicles Circ. Nucleic Acids 2024, 5, 64–82. [Google Scholar] [CrossRef]
- Palanisamy, C.P.; Pei, J.; Alugoju, P.; Anthikapalli, N.V.A.; Jayaraman, S.; Veeraraghavan, V.P.; Gopathy, S.; Roy, J.R.; Janaki, C.S.; Thalamati, D.; et al. New strategies of neurodegenerative disease treatment with extracellular vesicles (EVs) derived from mesenchymal stem cells (MSCs). Theranostics 2023, 13, 4138–4165. [Google Scholar] [CrossRef]
- Ma, Y.; Dong, S.; Li, X.; Kim, B.Y.S.; Yang, Z.; Jiang, W. Extracellular Vesicles: An Emerging Nanoplatform for Cancer Therapy. Front. Oncol. 2021, 10, 606906. [Google Scholar] [CrossRef]
- Weber, B.; Ritter, A.; Han, J.; Schaible, I.; Sturm, R.; Relja, B.; Huber-Lang, M.; Hildebrand, F.; Pallas, C.; Widera, M.; et al. Development of a Sampling and Storage Protocol of Extracellular Vesicles (EVs)—Establishment of the First EV Biobank for Polytraumatized Patients. Int. J. Mol. Sci. 2024, 25, 5645. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Andreu, Z.; Rivas, E.; Sanguino-Pascual, A.; Lamana, A.; Marazuela, M.; González-Alvaro, I.; Sánchez-Madrid, F.; de la Fuente, H.; Yáñez-Mó, M. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles 2016, 5, 31655. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lorite, P.; Domínguez, J.N.; Palomeque, T.; Torres, M.I. Extracellular Vesicles: Advanced Tools for Disease Diagnosis, Monitoring, and Therapies. Int. J. Mol. Sci. 2024, 26, 189. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ter-Ovanesyan, D.; Norman, M.; Lazarovits, R.; Trieu, W.; Lee, J.-H.; Church, G.M.; Walt, D.R. Framework for rapid comparison of extracellular vesicle isolation methods. eLife 2021, 10, e70725. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef] [PubMed]
- Dilsiz, N. A comprehensive review on recent advances in exosome isolation and characterization: Toward clinical applications. Transl. Oncol. 2024, 50, 102121. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- De Sousa, K.P.; Rossi, I.; Abdullahi, M.; Ramirez, M.I.; Stratton, D.; Inal, J.M. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1835. [Google Scholar] [CrossRef]
- de Araùjo, M.E.; Huber, L.A.; Stasyk, T. Isolation of endocitic organelles by density gradient centrifugation. Methods Mol. Biol. 2008, 424, 317–331. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. JCS 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.J.; de Oliveira, G.P., Jr.; Su, X.; Wood, J.; Fu, Z.; Pinckney, B.; Tigges, J.; Ghiran, I.; Ivanov, A.R. Multimode chromatography-based techniques for high purity isolation of extracellular vesicles from human blood plasma. J. Extracell. Biol. 2024, 3, e147. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, X.; Liang, J.; Ji, Y.; Zhou, Y.; Fang, H. Preparation of Silk Fibroin/Carboxymethyl Chitosan Hydrogel under Low Voltage as a Wound Dressing. Int. J. Mol. Sci. 2021, 22, 7610. [Google Scholar] [CrossRef]
- Nigro, A.; Finardi, A.; Ferraro, M.M.; Manno, D.E.; Quattrini, A.; Furlan, R.; Romano, A. Selective loss of microvesicles is a major issue of the differential centrifugation isolation protocols. Sci. Rep. 2021, 11, 3589. [Google Scholar] [CrossRef]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef]
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, E.K.; Yim, J.; Lee, M.H.; Lee, E.; Lee, Y.S.; Seo, W. Exosomes: Nomenclature, Isolation, and Biological Roles in Liver Diseases. Biomol. Ther. 2023, 31, 253–263. [Google Scholar] [CrossRef]
- Deregibus, M.C.; Figliolini, F.; D’Antico, S.; Manzini, P.M.; Pasquino, C.; De Lena, M.; Tetta, C.; Brizzi, M.F.; Camussi, G. Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 2016, 38, 1359–1366. [Google Scholar] [CrossRef]
- ExoQuick™ Exosome Precipitation Solution Cat# EXOQ5A-1 Cat# EXOQ20A-1 User Manual System Bioscience SBI. Available online: https://www.systembio.com/wp/wp-content/uploads/MANUAL_EXOQXXA-1-1.pdf (accessed on 16 January 2025).
- Yakubovich, E.I.; Polischouk, A.G.; Evtushenko, V.I. Principles and Problems of Exosome Isolation from Biological Fluids. Biochem. (Mosc.) Suppl. Ser. A Membr. Cell Biol. 2022, 16, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, N.; Hobiger, S.; Jungbauer, A. Continuous polyethylene glycol precipitation of recombinant antibodies: Sequential precipitation and resolubilization. Process Biochem. 2016, 51, 325–332. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Kastelowitz, N.; Yin, H. Exosomes and microvesicles: Identification and targeting by particle size and lipid chemical probes. ChemBioChem 2014, 15, 923–928. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Y.; Liu, Y.F.; Liu, H.Y.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Smolarz, M.; Pietrowska, M.; Matysiak, N.; Mielańczyk, Ł.; Widłak, P. Proteome profiling of exosomes purified from a small amount of human serum: The problem of co-purified serum components. Proteomes 2019, 7, 18. [Google Scholar] [CrossRef]
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; McCague, S.B.; Munro, B.R.; Predebon, M.J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018, 17, 211. [Google Scholar] [CrossRef]
- Wan, Z.; Zhao, L.; Lu, F.; Gao, X.; Dong, Y.; Zhao, Y.; Wei, M.; Yang, G.; Xing, C.; Liu, L. Mononuclear phagocyte system blockade improves therapeutic exosome delivery to the myocardium. Theranostics 2020, 10, 218–230. [Google Scholar] [CrossRef]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Su, C. Design strategies and application progress of therapeutic exosomes. Theranostics 2019, 9, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Deng, C.X. Current progresses of exosomes as cancer diagnostic and prognostic biomarkers. Int. J. Biol. Sci. 2019, 15, 1–11. [Google Scholar] [CrossRef]
- Rupp, A.K.; Rupp, C.; Keller, S.; Brase, J.C.; Ehehalt, R.; Fogel, M.; Moldenhauer, G.; Marmé, F.; Sültmann, H.; Altevogt, P. Loss of EpCAM expression in breast cancer-derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011, 122, 437–446. [Google Scholar] [CrossRef]
- Sankpal, N.V.; Brown, T.C.; Fleming, T.P.; Herndon, J.M.; Amaravati, A.A.; Loynd, A.N.; Gillanders, W.E. Cancer-associated mutations reveal a novel role for EpCAM as an inhibitor of cathepsin-L and tumor cell invasion. BMC Cancer 2021, 21, 541. [Google Scholar] [CrossRef]
- Wu, S.; Zhao, Y.; Zhang, Z.; Zuo, C.; Wu, H.; Liu, Y. The Advances and Applications of Characterization Technique for Exosomes: From Dynamic Light Scattering to Super-Resolution Imaging Technology. Photonics 2024, 11, 101. [Google Scholar] [CrossRef]
- Xian, P.; Hei, Y.; Wang, R.; Wang, T.; Yang, J.; Li, J.; Di, Z.; Liu, Z.; Baskys, A.; Liu, W.; et al. Mesenchymal stem cell-derived exosomes as a nanotherapeutic agent for amelioration of inflammation-induced astrocyte alterations in mice. Theranostics 2019, 9, 5956–5975. [Google Scholar] [CrossRef]
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935. [Google Scholar] [CrossRef]
- Lucchetti, D.; Battaglia, A.; Ricciardi-Tenore, C.; Colella, F.; Perelli, L.; De Maria, R.; Scambia, G.; Sgambato, A.; Fattorossi, A. Measuring Extracellular Vesicles by Conventional Flow Cytometry: Dream or Reality? Int. J. Mol. Sci. 2020, 21, 6257. [Google Scholar] [CrossRef]
- Erdbrügger, U.; Lannigan, J. Analytical challenges of extracellular vesicle detection: A comparison of different techniques. Cytom. Part A 2016, 89, 123–134. [Google Scholar] [CrossRef]
- de Rond, L.; van der Pol, E.; Bloemen, P.R.; Van Den Broeck, T.; Monheim, L.; Nieuwland, R.; van Leeuwen, T.G.; Coumans, F.A.W. A Systematic Approach to Improve Scatter Sensitivity of a Flow Cytometer for Detection of Extracellular Vesicles. Cytom. Part A 2020, 97, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Shen, H.; Li, Y.; Xing, Y.; Wang, J.; Guo, C.; Huang, Y.; Chen, J. Optimization of Flow Cytometric Sorting Parameters for High-Throughput Isolation and Purification of Small Extracellular Vesicles. J. Vis. Exp. JoVE 2023, 191, 64360. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol. Ther. 2018, 26, 2838–2847. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dehghani, M.; Lucas, K.; Flax, J.; McGrath, J.; Gaborski, T. Tangential flow microfluidics for the capture and release of nanoparticles and extracellular vesicles on conventional and ultrathin membranes. Adv. Mater. Technol. 2019, 4, 1900539. [Google Scholar] [CrossRef] [PubMed]
- Busatto, S.; Vilanilam, G.; Ticer, T.; Lin, W.L.; Dickson, D.W.; Shapiro, S.; Bergese, P.; Wolfram, J. Tangential Flow Filtration for Highly Efficient Concentration of Extracellular Vesicles from Large Volumes of Fluid. Cells 2018, 7, 273. [Google Scholar] [CrossRef]
- Lai, J.J.; Chau, Z.L.; Chen, S.Y.; Hill, J.J.; Korpany, K.V.; Liang, N.W.; Lin, L.H.; Lin, Y.H.; Liu, J.K.; Liu, Y.C.; et al. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv. Sci. 2022, 9, e2103222. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sun, H.; Duan, H.; Sheng, G.; Tian, N.; Liu, D.; Sun, Z. Isolation and usage of exosomes in central nervous system diseases. CNS Neurosci. Ther. 2024, 30, e14677. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef]
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef]
- Lozano-Ramos, I.; Bancu, I.; Oliveira-Tercero, A.; Armengol, M.P.; Menezes-Neto, A.; Del Portillo, H.A.; Lauzurica-Valdemoros, R.; Borràs, F.E. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles 2015, 4, 27369. [Google Scholar] [CrossRef]
- Muller, L.; Hong, C.-S.; Stolz, D.B.; Watkins, S.C.; Whiteside, T.L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods 2014, 411, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Rood, I.M.; Deegens, J.K.; Merchant, M.L.; Tamboer, W.P.; Wilkey, D.W.; Wetzels, J.F.; Klein, J.B. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010, 78, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef]
- Abramowicz, A.; Widlak, P.; Pietrowska, M. Proteomic analysis of exosomal cargo: The challenge of high purity vesicle isolation. Mol. BioSyst. 2016, 12, 1407–1419. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef]
- Welton, J.L.; Webber, J.P.; Botos, L.-A.; Jones, M.; Clayton, A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J. Extracell. Vesicles 2015, 4, 27269. [Google Scholar] [CrossRef]
- Darabi, S.; Ariaei, A.; Rustamzadeh, A.; Afshari, D.; Charkhat Gorgich, E.A.; Darabi, L. Cerebrospinal fluid and blood exosomes as biomarkers for amyotrophic lateral sclerosis; a systematic review. Diagn. Pathol. 2024, 19, 47. [Google Scholar] [CrossRef]
- de Menezes-Neto, A.; Sáez, M.J.; Lozano-Ramos, I.; Segui-Barber, J.; Martin-Jaular, L.; Ullate, J.M.; Fernandez-Becerra, C.; Borrás, F.E.; Del Portillo, H.A. Size-exclusion chromatography as a stand-alone methodology identifies novel markers in mass spectrometry analyses of plasma-derived vesicles from healthy individuals. J. Extracell. Vesicles 2015, 4, 27378. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dudani, J.S.; Gossett, D.R.; Tse, H.T.K.; Lamm, R.J.; Kulkarni, R.P.; Di Carlo, D. Rapid inertial solution exchange for enrichment and flow cytometric detection of microvesicles. Biomicrofluidics 2015, 9, 014112. [Google Scholar] [CrossRef]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small 2020, 16, 1903916. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, S.; Shehata Draz, M.; Zarghooni, M.; Sanati-Nezhad, A.; Ghavami, S.; Shafiee, H.; Akbari, M. Microfluidic approaches for isolation, detection, and characterization of extracellular vesicles: Current status and future directions. Biosens. Bioelectron. 2017, 91, 588–605. [Google Scholar] [CrossRef] [PubMed]
- Narayanamurthy, V.; Jeroish, Z.E.; Bhuvaneshwari, K.S.; Bayat, P.; Premkumar, R.; Samsuri, F.; Yusoff, M.M. Advances in passively driven microfluidics and lab-on-chip devices: A comprehensive literature review and patent analysis. RSC Adv. 2020, 10, 11652–11680. [Google Scholar] [CrossRef]
- Guo, W.; Gao, Y.; Li, N.; Shao, F.; Wang, C.; Wang, P.; Yang, Z.; Li, R.; He, J. Exosomes: New players in cancer. Oncol. Rep. 2017, 38, 665–675. [Google Scholar] [CrossRef]
- Oliveira-Rodríguez, M.; López-Cobo, S.; Reyburn, H.T.; Costa-García, A.; López-Martín, S.; Yáñez-Mó, M.; Cernuda-Morollón, E.; Paschen, A.; Valés-Gómez, M.; Blanco-López, M.C. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J. Extracell. Vesicle 2016, 5, 31803. [Google Scholar] [CrossRef]
- Zhao, Z.; Wijerathne, H.; Godwin, A.K.; Soper, S.A. Isolation and analysis methods of extracellular vesicles (EVs). Extracell. Vesicles Circ. Nucl. Acids 2021, 2, 80–103. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, H.J.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.; Liu, X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip 2013, 13, 2879. [Google Scholar] [CrossRef]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Xue, Y.; Qiao, L.; Yu, G.; Liu, Y.; Yu, S. Ultrasensitive analysis of exosomes using a 3D self-assembled nanostructured SiO2 microfluidic chip. ACS Appl. Mater. Interfaces 2022, 14, 14693–14702. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, Y.; Bühler, M.; Wang, S.; Asghari, M.; Stürchler, A.; Mateescu, B.; Weiss, T.; Stavrakis, S.; deMello, A.J. Direct isolation of small extracellular vesicles from human blood using viscoelastic microfluidics. Sci. Adv. 2023, 9, eadi5296. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Liao, X.; Tian, Y.; Li, G. Exosome separation using microfluidic systems: Size-based, immunoaffinity-based and dynamic methodologies. Biotechnol. J. 2017, 12, 1600699. [Google Scholar] [CrossRef] [PubMed]
- Mogi, K.; Hayashida, K.; Yamamoto, T. Damage-less handling of exosomes using an ion-depletion zone in a microchannel. Anal. Sci. 2018, 34, 875–880. [Google Scholar] [CrossRef]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef]
- Midekessa, G.; Godakumara, K.; Ord, J.; Viil, J.; Lättekivi, F.; Dissanayake, K.; Kopanchuk, S.; Rinken, A.; Andronowska, A.; Bhattacharjee, S.; et al. Zeta Potential of Extracellular Vesicles: Toward Understanding the Attributes that Determine Colloidal Stability. ACS Omega 2020, 5, 16701–16710. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mendivil-Alvarado, H.; Limon-Miro, A.T.; Carvajal-Millan, E.; Lizardi-Mendoza, J.; Mercado-Lara, A.; Coronado-Alvarado, C.D.; Rascón-Durán, M.L.; Anduro-Corona, I.; Talamás-Lara, D.; Rascón-Careaga, A.; et al. Extracellular Vesicles and Their Zeta Potential as Future Markers Associated with Nutrition and Molecular Biomarkers in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 6810. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guerrero-Alba, A.; Bansal, S.; Sankpal, A.N.; Mitra, G.; Rahman, M.; Ravichandran, R.; Poulson, C.; Fleming, T.P.; Smith, M.A.; Bremner, R.M.; et al. Enhanced enrichment of extracellular vesicles for laboratory and clinical research from drop-sized blood samples. Front. Mol. Biosci. 2024, 11, 1365783. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Welsh, J.A.; Arkesteijn, G.J.A.; Bremer, M.; Cimorelli, M.; Dignat-George, F.; Giebel, B.; Görgens, A.; Hendrix, A.; Kuiper, M.; Lacroix, R.; et al. A compendium of single extracellular vesicle flow cytometry. J. Extracell. Vesicles 2023, 12, e12299. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pleet, M.L.; Cook, S.; Tang, V.A.; Stack, E.; Ford, V.J.; Lannigan, J.; Do, N.; Wenger, E.; Fraikin, J.L.; Jacobson, S.; et al. Extracellular Vesicle Refractive Index Derivation Utilizing Orthogonal Characterization. Nano Lett. 2023, 23, 9195–9202. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van der Pol, E.; Coumans, F.A.; Grootemaat, A.E.; Gardiner, C.; Sargent, I.L.; Harrison, P.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemost. 2014, 12, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.J.; Kaur, H.; Dobhal, G.; Malhotra, S.; Ayed, Z.; Carpenter, A.L.; Goreham, R.V. The Physical Characterization of Extracellular Vesicles for Function Elucidation and Biomedical Applications: A Review. Part. Part. Syst. Charact. 2024, 41, 2400024. [Google Scholar] [CrossRef]
- Askeland, A.; Borup, A.; Østergaard, O.; Olsen, J.V.; Lund, S.M.; Christiansen, G.; Kristensen, S.R.; Heegaard, N.H.H.; Pedersen, S. Mass-Spectrometry Based Proteome Comparison of Extracellular Vesicle Isolation Methods: Comparison of ME-kit, Size-Exclusion Chromatography, and High-Speed Centrifugation. Biomedicines 2020, 8, 246. [Google Scholar] [CrossRef]
- Manouchehri Doulabi, E.; Fredolini, C.; Gallini, R.; Löf, L.; Shen, Q.; Ikebuchi, R.; Dubois, L.; Azimi, A.; Loudig, O.; Gabrielsson, S.; et al. Surface protein profiling of prostate-derived extracellular vesicles by mass spectrometry and proximity assays. Commun. Biol. 2022, 5, 1402. [Google Scholar] [CrossRef]
- Kowkabany, G.; Bao, Y. Nanoparticle Tracking Analysis: An Effective Tool to Characterize Extracellular Vesicles. Molecules 2024, 29, 4672. [Google Scholar] [CrossRef]
- Kang, D.; Oh, S.; Ahn, S.M.; Lee, B.H.; Moon, M.H. Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation and nanoflow liquid chromatography–tandem mass spectrometry. J. Proteome Res. 2008, 7, 3475–3480. [Google Scholar] [CrossRef]
- Ouyang, W.; Aristov, A.; Lelek, M.; Hao, X.; Zimmer, C. Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 2018, 36, 460–468. [Google Scholar] [CrossRef]
- Bonilla, H.; Hampton, D.; Marques de Menezes, E.G.; Deng, X.; Montoya, J.G.; Anderson, J.; Norris, P.J. Comparative Analysis of Extracellular Vesicles in Patients with Severe and Mild Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front. Immunol. 2022, 13, 841910. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kurtjak, M.; Kereïche, S.; Klepac, D.; Križan, H.; Perčić, M.; Krušić Alić, V.; Lavrin, T.; Lenassi, M.; Wechtersbach, K.; Kojc, N.; et al. Unveiling the Native Morphology of Extracellular Vesicles from Human Cerebrospinal Fluid by Atomic Force and Cryogenic Electron Microscopy. Biomedicines 2022, 10, 1251. [Google Scholar] [CrossRef]
- Parisse, P.; Rago, I.; Ulloa Severino, L.; Perissinotto, F.; Ambrosetti, E.; Paoletti, P.; Ricci, M.; Beltrami, A.P.; Cesselli, D.; Casalis, L. Atomic force microscopy analysis of extracellular vesicles. Eur. Biophys. J. 2017, 46, 813–820. [Google Scholar] [CrossRef]
- Sharma, S.; LeClaire, M.; Gimzewski, J.K. Ascent of atomic force microscopy as a nanoanalytical tool for exosomes and other extracellular vesicles. Nanotechnology 2018, 29, 132001. [Google Scholar] [CrossRef]
- Datta, A.; Kim, H.; McGee, L.; Johnson, A.E.; Talwar, S.; Marugan, J.; Southall, N.; Hu, X.; Lal, M.; Mondal, D.; et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci. Rep. 2018, 8, 8161. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coumans, F.A.; van der Pol, E.; Böing, A.N.; Hajji, N.; Sturk, G.; van Leeuwen, T.G.; Nieuwland, R. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J. Extracell. Vesicles 2014, 3, 25922. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhu, L.; Zhang, W.; Elnatan, D.; Huang, B. Faster STORM using compressed sensing. Nat. Methods 2012, 9, 721–723. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Gong, W.; Yang, Z.; Pan, W.; Verwilst, P.; Shin, J.; Yan, W.; Liu, L.; Qu, J.; Kim, J.S. STORM imaging of mitochondrial dynamics using a vicinal-dithiol-proteins-targeted probe. Biomaterials 2020, 243, 119938. [Google Scholar] [CrossRef] [PubMed]
- Helmerich, D.A.; Beliu, G.; Taban, D.; Meub, M.; Streit, M.; Kuhlemann, A.; Doose, S.; Sauer, M. Photoswitching fingerprint analysis bypasses the 10-nm resolution barrier. Nat. Methods 2022, 19, 986–994. [Google Scholar] [CrossRef]
- Dechantsreiter, S.; Ambrose, A.R.; Worboys, J.D.; Lim, J.M.E.; Liu, S.; Shah, R.; Montero, M.A.; Quinn, A.M.; Hussell, T.; Tannahill, G.M.; et al. Heterogeneity in extracellular vesicle secretion by single human macrophages revealed by super-resolution microscopy. J. Extracell. Vesicles 2022, 11, e12215. [Google Scholar] [CrossRef]
- Stepanenko, T.; Sofińska, K.; Wilkosz, N.; Dybas, J.; Wiercigroch, E.; Bulat, K.; Szczesny-Malysiak, E.; Skirlińska-Nosek, K.; Seweryn, S.; Chwiej, J.; et al. Surface-enhanced Raman scattering (SERS) and tip-enhanced Raman scattering (TERS) in label-free characterization of erythrocyte membranes and extracellular vesicles at the nano-scale and molecular level. Analyst 2024, 149, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Buccini, L.; Proietti, A.; La Penna, G.; Mancini, C.; Mura, F.; Tacconi, S.; Dini, L.; Rossi, M.; Passeri, D. Toward the nanoscale chemical and physical probing of milk-derived extracellular vesicles using Raman and tip-enhanced Raman spectroscopy. Nanoscale 2024, 16, 8132–8142. [Google Scholar] [CrossRef] [PubMed]
- Raizada, G.; Brunel, B.; Guillouzouic, J.; Aubertin, K.; Shigeto, S.; Nishigaki, Y.; Lesniewska, E.; Le Ferrec, E.; Boireau, W.; Elie-Caille, C. Raman spectroscopy of large extracellular vesicles derived from human microvascular endothelial cells to detect benzo[a]pyrene exposure. Anal. Bioanal. Chem. 2024, 416, 6639–6649. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.M.; Sida, L.A.; Tűzesi, C.Á.; Shivalingam, B.; Sim, H.W.; Buckland, M.E.; Satgunaseelan, L.; Alexander, K.L. Size matters: Biomolecular compositions of small and large extracellular vesicles in the urine of glioblastoma patients. J. Extracell. Biol. 2024, 3, e70021, Erratum in: J. Extracell. Biol. 2024, 3, e70026. https://doi.org/10.1002/jex2.70026. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bajaj, N.; Sharma, D. Uncovering metabolic signatures in cancer-derived exosomes: LC-MS/MS and NMR profiling. Nanoscale 2024, 17, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Vaz, M.; Soares Martins, T.; Leandro, K.; de Almeida, L.P.; da Cruz e Silva, O.A.B.; Nunes, A.; Henriques, A.G. Fourier Transform Infrared Spectroscopy Analysis as a Tool to Address Aβ Impact on Extracellular Vesicles. Molecules 2025, 30, 258. [Google Scholar] [CrossRef] [PubMed]
- Starchev, K.; Buffle, J.; Pérez, E. Applications of fluorescence correlation spectroscopy: Polydispersity measurements. J. Colloid Interface Sci. 1999, 213, 479–487. [Google Scholar] [CrossRef]
- Vidal, M.; Mangeat, P.; Hoekstra, D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J. Cell Sci. 1997, 110, 1867–1877. [Google Scholar] [CrossRef]
- Van der Pol, E. Methods for detection and characterization of extracellular vesicles. In Detection of Extracellular Vesicles: Size Does Matter; Uitgeverij Box Press: Vianen, The Netherlands, 2015; pp. 29–47. [Google Scholar]
- Kirz, J.; Jacobsen, C. Soft X-ray microscopes and their biological applications List of Figures. Q. Rev. Biophys. 1995, 28, 33–130. [Google Scholar] [CrossRef]
- Logozzi, M.; de Milito, A.; Lugini, L.; Borghi, M.; Calabrò, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Kosaka, N.; Konishi, Y.; Ohta, H.; Okamoto, H.; Sonoda, H.; Nonaka, R.; Yamamoto, H.; Ishii, H.; Mori, M.; et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014, 5, 3591. [Google Scholar] [CrossRef]
- Mizenko, R.R.; Brostoff, T.; Rojalin, T.; Koster, H.J.; Swindell, H.S.; Leiserowitz, G.S.; Wang, A.; Carney, R.P. Tetraspanins are unevenly distributed across single extracellular vesicles and bias sensitivity to multiplexed cancer biomarkers. J. Nanobiotechnol. 2021, 19, 250. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Sessa, F.; Salerno, M.; Esposito, M.; Cocimano, G.; Pomara, C. miRNA Dysregulation in Cardiovascular Diseases: Current Opinion and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 5192. [Google Scholar] [CrossRef]
- Quang, M.T.; Nguyen, M.N. The potential of microRNAs in cancer diagnostic and therapeutic strategies: A narrative review. JoBAZ 2024, 85, 7. [Google Scholar] [CrossRef]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Bergmans, B.; Papadopoulou, A.S.; Delacourte, A.; De Strooper, B. MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol Dis. 2009, 33, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Michlewski, G.; Guil, S.; Semple, C.A.; Cáceres, J.F. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008, 32, 383–393. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chang, T.C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007, 26, 745–752. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009, 459, 1010–1014. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Blazie, S.M.; Geissel, H.C.; Wilky, H.; Joshi, R.; Newbern, J.; Mangone, M. Alternative Polyadenylation Directs Tissue-Specific miRNA Targeting in Caenorhabditis elegans Somatic Tissues. Genetics 2017, 206, 757–774. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hoque, P.; Romero, B.; Akins, R.E.; Batish, M. Exploring the Multifaceted Biologically Relevant Roles of circRNAs: From Regulation, Translation to Biomarkers. Cells 2023, 12, 2813. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, X.; Zuo, X.; Yang, B.; Li, Z.; Xue, Y.; Zhou, Y.; Huang, J.; Zhao, X.; Zhou, J.; Yan, Y.; et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014, 158, 607–619. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Gál, L.; Schamberger, A.; Wachtl, G.; Orbán, T.I. The Effect of Alternative Splicing Sites on Mirtron Formation and Arm Selection of Precursor microRNAs. Int J Mol Sci. 2024, 25, 7643. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abolhasani, S.; Ahmadi, Y.; Rostami, Y.; Fattahi, D. The role of MicroRNAs in mesenchymal stem cell differentiation into vascular smooth muscle cells. Cell Div. 2025, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Azizan, A.; Farhadi, E.; Faezi, S.T.; Jamshidi, A.; Alikhani, M.; Mahmoudi, M. Role of miRNAs in Apoptosis Pathways of Immune Cells in Systemic Lupus Erythematosus. Immun. Inflamm. Dis. 2025, 13, e70124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bagni, G.; Biancalana, E.; Chiara, E.; Costanzo, I.; Malandrino, D.; Lastraioli, E.; Palmerini, M.; Silvestri, E.; Urban, M.L.; Emmi, G. Epigenetics in autoimmune diseases: Unraveling the hidden regulators of immune dysregulation. Autoimmun. Rev. 2025, 24, 103784. [Google Scholar] [CrossRef] [PubMed]
- Gaál, Z. Role of microRNAs in Immune Regulation with Translational and Clinical Applications. Int. J. Mol. Sci. 2024, 25, 1942. [Google Scholar] [CrossRef] [PubMed]
- Masliah-Planchon, J.; Garinet, S.; Pasmant, E. RAS-MAPK pathway epigenetic activation in cancer: miRNAs in action. Oncotarget 2016, 7, 38892–38907. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, T.; Croce, C.M. MicroRNA: Trends in clinical trials of cancer diagnosis and therapy strategies. Exp Mol Med. 2023, 1314–1321. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Junn, E.; Mouradian, M.M. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther. 2012, 133, 142–150. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Singh, V.; Sen, A.; Saini, S.; Dwivedi, S.; Agrawal, R.; Bansal, A.; Shekhar, S. MicroRNA Significance in Cancer: An Updated Review on Diagnostic, Prognostic, and Therapeutic Perspectives. EJIFCC 2024, 35, 265–284. [Google Scholar]
- Miroshnichenko, S.; Patutina, O. Enhanced Inhibition of Tumorigenesis Using Combinations of miRNA-Targeted Therapeutics. Front. Pharmacol. 2019, 10, 488. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Liu, R.; Lv, X.; He, H.; Song, D.; Chen, Y.; Zheng, C.; Lin, Y.-F.; Xu, Q.; He, R.-L.; et al. Antagomir-21 Improve Post-MI Heart Failure by Inhibiting Myocardial Fibrosis and Myocardial Apoptosis. Available online: https://ssrn.com/abstract=4385200 (accessed on 2 January 2025).
- Huang, X.; Li, S.; Qiu, N.; Ni, A.; Xiong, T.; Xue, J.; Yin, K.J. Sex and Age-Dependent Effects of miR-15a/16-1 Antagomir on Ischemic Stroke Outcomes. Int. J. Mol. Sci. 2024, 25, 11765. [Google Scholar] [CrossRef]
- Razaviyan, J.; Sirati-Sabet, M.; Tafti, A.; Hadavi, R.; Karima, S.; Rajabibazl, M.; Mohammadi-Yeganeh, S. Inhibition of MiR-155 Using Exosomal Delivery of Antagomir Can Up-Regulate PTEN in Triple Negative Breast Cancer. Endocr. Metab. Immune Disord. Drug Targets 2024, 24, 1664–1676. [Google Scholar] [CrossRef]
- Nguyen, L.D.; Wei, Z.; Silva, M.C.; Barberán-Soler, S.; Zhang, J.; Rabinovsky, R.; Muratore, C.R.; Stricker, J.M.S.; Hortman, C.; Young-Pearse, T.L.; et al. Small molecule regulators of microRNAs identified by high-throughput screen coupled with high-throughput sequencing. Nat. Commun. 2023, 14, 7575. [Google Scholar] [CrossRef]
- Panigrahi, M.; Palmer, M.A.; Wilson, J.A. MicroRNA-122 Regulation of HCV Infections: Insights from Studies of miR-122-Independent Replication. Pathogens 2022, 11, 1005. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, M.; Thibault, P.A.; Wilson, J.A. MicroRNA 122 Affects both the Initiation and the Maintenance of Hepatitis C Virus Infections. J. Virol. 2022, 96, e0190321. [Google Scholar] [CrossRef] [PubMed]
- Emanuelson, C.; Ankenbruck, N.; Kumbhare, R.; Thomas, M.; Connelly, C.; Baktash, Y.; Randall, G.; Deiters, A. Transcriptional Inhibition of MicroRNA miR-122 by Small Molecules Reduces Hepatitis C Virus Replication in Liver Cells. J. Med. Chem. 2022, 65, 16338–16352. [Google Scholar] [CrossRef] [PubMed]
- Quilón, P.G.; Volpedo, G.; Cappato, S.; Ferrera, L.; Zara, F.; Bocciardi, R.; Riva, A.; Striano, P. Antisense oligonucleotides as a precision therapy for developmental and epileptic encephalopathies. CNS Neurosci. Ther. 2024, 30, e70050. [Google Scholar] [CrossRef]
- Hong, L.Z.; Zhou, L.; Zou, R.; Khoo, C.M.; Chew, A.L.S.; Chin, C.L.; Shih, S.J. Systematic evaluation of multiple qPCR platforms, NanoString, and miRNA-Seq for microRNA biomarker discovery in human biofluids. Sci. Rep. 2021, 11, 4435. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef]
- Crawford, M.; Brawner, E.; Batte, K.; Yu, L.; Hunter, M.G.; Otterson, G.A.; Nuovo, G.; Marsh, C.B.; Nana-Sinkam, S.P. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun. 2009, 373, 607–612. [Google Scholar] [CrossRef]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef]
- BMC Infectious Diseases. The potential of circulating microRNAs as novel diagnostic biomarkers in COVID-19. BMC Infect. Dis. 2024, 24, 9915. [Google Scholar]
- Kong, D.; Wang, K.; Zhang, Q.-N.; Bing, Z.-T. Systematic analysis reveals key microRNAs as diagnostic and prognostic factors in progressive stages of lung cancer. arXiv 2022, arXiv:2201.05408. [Google Scholar]
- Pimalai, D.; Putnin, T.; Waiwinya, W.; Aroonyadet, N. Development of electrochemical biosensors for simultaneous multiplex detection of microRNA for breast cancer screening. Microchim. Acta 2021, 188, 314. [Google Scholar] [CrossRef] [PubMed]
- Hilario, E. End labeling procedures: An overview. Mol. Biotechnol. 2004, 28, 77–80. [Google Scholar] [CrossRef]
- Amanat, M.; Nemeth, C.L.; Fine, A.S.; Leung, D.G.; Fatemi, A. Antisense Oligonucleotide Therapy for the Nervous System: From Bench to Bedside with Emphasis on Pediatric Neurology. Pharmaceutics 2022, 14, 2389. [Google Scholar] [CrossRef]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef]
- Tijsen, A.J.; Pinto, Y.M.; Creemers, E.E. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H1085–H1095. [Google Scholar] [CrossRef]
- Romaine, S.P.; Tomaszewski, M.; Condorelli, G.; Samani, N.J. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart 2015, 101, 921–928. [Google Scholar] [CrossRef]
- Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef]
- Peng, J.; Hang, X.; Wang, H.; Zhao, K.; Wang, H.; Yu, H.; Wang, L. Low background self-primer EXPAR coupled with colorimetric and lateral flow assay for rapid and sensitive point-of-care detection of miRNA. Sens. Actuators B Chem. 2024, 399, 134856. [Google Scholar] [CrossRef]
- de Ronde, M.W.J.; Ruijter, J.M.; Lanfear, D.; Bayes-Genis, A.; Kok, M.G.M.; Creemers, E.E.; Pinto, Y.M.; Pinto-Sietsma, S.J. Practical data handling pipeline improves performance of qPCR-based circulating miRNA measurements. RNA 2017, 23, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological Challenges in Utilizing miRNAs as Circulating Biomarkers. J. Cell Mol. Med. 2014, 18, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Xiao, S.; Gao, Y.; Soung, Y.H. Recent Technologies Towards Diagnostic and Therapeutic Applications of Circulating Nucleic Acids in Colorectal Cancers. Int. J. Mol. Sci. 2024, 25, 8703. [Google Scholar] [CrossRef]
- Zampetaki, A.; Mayr, M. Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb. Haemost. 2012, 108, 592–598. [Google Scholar]
- Gibson, U.E.; Heid, C.A.; Williams, P.M. A novel method for real-time quantitative RT-PCR. Genome Res. 1996, 6, 995–1001. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Pfaffl, M.W.; Zhao, S.; Spiess, A.N.; Boggy, G.; Blom, J.; Rutledge, R.G.; Sisti, D.; Lievens, A.; De Preter, K.; et al. Evaluation of qPCR curve analysis methods for reliable biomarker discovery: Bias, resolution, precision, and implications. Methods 2013, 59, 32–46. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Abdullah Al-Maskri, A.A.; Ye, J.; Talap, J.; Hu, H.; Sun, L.; Yu, L.; Cai, S.; Zeng, S. Reverse transcription-based loop-mediated isothermal amplification strategy for real-time miRNA detection with phosphorothioated probes. Anal. Chim. Acta 2020, 1126, 1–6. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010, 16, 991–1006. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Diehl, P.; Fricke, A.; Sander, L.; Stamm, J.; Bassler, N.; Htun, N.; Ziemann, M.; Helbing, T.; El-Osta, A.; Jowett, J.B.; et al. Microparticles: Major transport vehicles for distinct microRNAs in circulation. Cardiovasc. Res. 2012, 93, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Borchert, G.M.; Dou, D.; Huan, J.; Lan, W.; Tan, M.; Wu, B. Bioinformatics in MicroRNA Research; Humana Press: New York, NY, USA, 2017. [Google Scholar]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Investig. 2019, 99, 452–469. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Baldwin, D.A.; Kloosterman, W.P.; Kauppinen, S.; Plasterk, R.H.; Mourelatos, Z. RAKE and LNA-ISH reveal microRNA expression and localization in RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 2006, 12, 187–191. [Google Scholar] [CrossRef] [PubMed]
- McDougall, J.K.; Dunn, A.R.; Jones, K.W. In situ hybridization of adenovirus RNA and DNA. Nature 1972, 236, 346–348. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Wienholds, E.; de Bruijn, E.; Kauppinen, S.; Plasterk, R.H. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods 2006, 3, 27–29. [Google Scholar] [CrossRef]
- Hanna, J.A.; Wimberly, H.; Kumar, S.; Slack, F.; Agarwal, S.; Rimm, D.L. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. Biotechniques 2012, 52, 235–245. [Google Scholar] [CrossRef]
- Parisi, F.; Micsinai, M.; Strino, F.; Ariyan, S.; Narayan, D.; Bacchiocchi, A.; Cheng, E.; Xu, F.; Li, P.; Kluger, H.; et al. Integrated analysis of tumor samples sheds light on tumor heterogeneity. Yale J. Biol. Med. 2012, 85, 347–361. [Google Scholar]
- Wei, Y.; Xu, F.; Li, P. Technology-driven and evidence-based genomic analysis for integrated pediatric and prenatal genetics evaluation. J. Genet. Genom. 2013, 40, 1–14. [Google Scholar] [CrossRef]
- Martin, C.L.; Warburton, D. Detection of chromosomal aberrations in clinical practice: From karyotype to genome sequence. Annu. Rev. Genom. Hum. Genet. 2015, 16, 309–326. [Google Scholar] [CrossRef]
- Tang, G.; Reinhart, B.J.; Bartel, D.P.; Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003, 17, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Böhm-Hofstätter, H.; Tschernutter, M.; Kunert, R. Comparison of hybridization methods and real-time PCR: Their value in animal cell line characterization. Appl. Microbiol. Biotechnol. 2010, 87, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Reue, K. mRNA quantitation techniques: Considerations for experimental design and application. J. Nutr. 1998, 128, 2038–2044. [Google Scholar] [CrossRef]
- Novogrodsky, A.; Hurwitz, J. The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid. I. Phosphorylation at 5’-hydroxyl termini. J. Biol. Chem. 1966, 241, 2933–2943. [Google Scholar] [CrossRef]
- Novara, C.; Montesi, D.; Bertone, S.; Paccotti, N.; Geobaldo, F.; Channab, M.; Angelini, A.; Rivolo, P.; Giorgis, F.; Chiadò, A. Label-free detection of miRNAs: Role of probe design and bioassay configuration in Surface Enhanced Raman Scattering based biosensors. arXiv 2023, arXiv:2301.07062. [Google Scholar]
- Yang, H.; Jin, Y.; Qian, H.; Wang, Y.; Bao, T.; Wu, Z.; Wen, W.; Zhang, X.; Wang, S. Target-driven cascade amplified assembly of covalent organic frameworks on tetrahedral DNA nanostructure with multiplex recognition domains for ultrasensitive detection of microRNA. Anal. Chim. Acta 2024, 1311, 342743. [Google Scholar] [CrossRef]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Moskovits, M. Surface roughness and the enhanced intensity of Raman scattering by molecules adsorbed on metals. J. Chem. Phys. 1978, 69, 4159–4161. [Google Scholar] [CrossRef]
- Osawa, M.; Matsuda, N.; Yoshii, K.; Uchida, I. Charge transfer resonance Raman process in surface-enhanced Raman scattering from p-aminothiophenol adsorbed on silver: Herzberg-Teller contribution. J. Phys. Chem. 1994, 98, 12702–12707. [Google Scholar] [CrossRef]
- Cong, S.; Liu, X.; Jiang, Y.; Zhang, W.; Zhao, Z. Surface enhanced Raman scattering revealed by interfacial charge-transfer transitions. Innovation 2020, 1, 100051. [Google Scholar] [CrossRef]
- Lombardi, J.R.; Birke, R.L. A unified view of surface-enhanced Raman scattering. Acc. Chem. Res. 2009, 42, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, J.R.; Birke, R.L. A unified approach to surface-enhanced Raman spectroscopy. J. Phys. Chem. 2008, 112, 5605–5617. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.; Huang, S.; Tai, J.; Wang, X.; Dai, X.; Qiu, C.; Gu, D.; Yuan, W.; Ho, H.P.; et al. Advancing MicroRNA Detection: Enhanced Biotin-Streptavidin Dual-Mode Phase Imaging Surface Plasmon Resonance Aptasensor. Anal. Chem. 2024, 96, 8791–8799. [Google Scholar] [CrossRef]
- Miotto, E.; Saccenti, E.; Lupini, L.; Callegari, E.; Negrini, M.; Ferracin, M. Quantification of circulating miRNAs by droplet digital PCR: Comparison of EvaGreen- and TaqMan-based chemistries. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 2638–2642. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Foye, C.; Yan, I.K.; David, W.; Shukla, N.; Habboush, Y.; Chase, L.; Ryland, K.; Kesari, V.; Patel, T. Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS ONE 2017, 12, e0189165. [Google Scholar] [CrossRef]
- Shang, Z.; Ding, D.; Deng, Z.; Zhao, J.; Yang, M.; Xiao, Y.; Chu, W.; Xu, S.; Zhang, Z.; Yi, X.; et al. Programming the Dynamic Range of Nanochannel Biosensors for MicroRNA Detection Through Allosteric DNA Probes. Angew. Chem. 2024, 137, e202417280. [Google Scholar] [CrossRef]
- Rouhi, S.; Ghasemi, H.; Alizadeh, M.; Movahedpour, A.; Vahedi, F.; Fattahi, M.; Aiiashi, S.; Khatami, S.H. miRNA-based electrochemical biosensors for ovarian cancer. Clin. Chim. Acta 2025, 564, 119946. [Google Scholar] [CrossRef]
- Sim, S.B.; Haizan, I.; Choi, M.Y.; Lee, Y.; Choi, J.-H. Recent Strategies for MicroRNA Detection: A Comprehensive Review of SERS-Based Nanobiosensors. Chemosensors 2024, 12, 154. [Google Scholar] [CrossRef]
- Ma, C.; Ding, R.; Hao, K.; Du, W.; Xu, L.; Gao, Q.; Yu, C. Storage Stability of Blood Samples for miRNAs in Glycosylated Extracellular Vesicles. Molecules 2024, 29, 103. [Google Scholar] [CrossRef]
- Akers, J.C.; Ramakrishnan, V.; Yang, I.; Hua, W.; Mao, Y.; Carter, B.S.; Chen, C.C. Optimizing preservation of extracellular vesicular miRNAs derived from clinical cerebrospinal fluid. Cancer Biomark. 2016, 17, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Di, K.; Fan, B.; Wu, J.; Gu, X.; Sun, Y.; Khan, A.; Li, P.; Li, Z. MicroRNAs in extracellular vesicles: Sorting mechanisms, diagnostic value, isolation, and detection technology. Front. Bioeng. Biotechnol. 2022, 10, 948959. [Google Scholar] [CrossRef] [PubMed]
- Llorens-Revull, M.; Martínez-González, B.; Quer, J.; Esteban, J.I.; Núñez-Moreno, G.; Mínguez, P.; Burgui, I.; Ramos-Ruíz, R.; Soria, M.E.; Rico, A.; et al. Comparison of Extracellular Vesicle Isolation Methods for miRNA Sequencing. Int. J. Mol. Sci. 2023, 24, 12183. [Google Scholar] [CrossRef]
- Bazrgar, M.; Khodabakhsh, P.; Dargahi, L.; Mohagheghi, F.; Ahmadiani, A. MicroRNA modulation is a potential molecular mechanism for neuroprotective effects of intranasal insulin administration in amyloid βeta oligomer induced Alzheimer’s like rat model. Exp. Gerontol. 2022, 164, 111812. [Google Scholar] [CrossRef]
- Hamid, Y.; Rabbani, R.D.; Afsara, R.; Nowrin, S.; Ghose, A.; Papadopoulos, V.; Sirlantzis, K.; Ovsepian, S.V.; Boussios, S. Exosomal Liquid Biopsy in Prostate Cancer: A Systematic Review of Biomarkers for Diagnosis, Prognosis, and Treatment Response. Int. J. Mol. Sci. 2025, 26, 802. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, Y.; Wang, Z.; Gao, Q.; Hao, K.; Chen, X.; Ke, N.; Lv, X.; Weng, J.; Zhong, Y.; et al. Development a glycosylated extracellular vesicle-derived miRNA Signature for early detection of esophageal squamous cell carcinoma. BMC Med. 2025, 23, 39. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Uratani, R.; Toiyama, Y.; Kitajima, T.; Kawamura, M.; Hiro, J.; Kobayashi, M.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; et al. Diagnostic Potential of Cell-Free and Exosomal MicroRNAs in the Identification of Patients with High-Risk Colorectal Adenomas. PLoS ONE 2016, 11, e0160722. [Google Scholar] [CrossRef]
- Machida, T.; Tomofuji, T.; Maruyama, T.; Yoneda, T.; Ekuni, D.; Azuma, T.; Miyai, H.; Mizuno, H.; Kato, H.; Tsutsumi, K.; et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol. Rep. 2016, 36, 2375–2381. [Google Scholar] [CrossRef]
- Khayamzadeh, M.; Niazi, V.; Hussen, B.M.; Taheri, M.; Ghafouri-Fard, S.; Samadian, M. Emerging role of extracellular vesicles in the pathogenesis of glioblastoma. Metab. Brain Dis. 2023, 38, 177–184. [Google Scholar] [CrossRef]
- Qiu, M.; Zhai, S.; Fu, Q.; Liu, D. Bone marrow mesenchymal stem cells-derived exosomal microRNA-150-3p promotes osteoblast proliferation and differentiation in osteoporosis. Hum. Gene Ther. 2021, 32, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-S.; Liu, J.-B.; Lin, L.; Zhang, H.; Wu, J.-J.; Shi, Y.; Jia, C.-Y.; Zhang, D.-D.; Yu, F.; Wang, H.-M. Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4. Cell Death Discov. 2021, 7, 224. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.; Yu, Y.J.; You, J.Y.; Rhee, W.J.; Kim, J.A. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip 2020, 20, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Ghodasara, A.; Raza, A.; Wolfram, J.; Salomon, C.; Popat, A. Clinical Translation of Extracellular Vesicles. Adv. Healthcare Mater. 2023, 12, e2301010. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.-M.; Yang, Q.-Y.; Monsel, A.; Yan, J.-Y.; Dai, C.-X.; Zhao, J.-Y.; Shi, G.-C.; Zhou, M.; Zhu, X.-M.; Li, S.-K.; et al. Preclinical efficacy and clinical safety of clinical-grade nebulized allogenic adipose mesenchymal stromal cells-derived extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12134. [Google Scholar] [CrossRef]
- Mizenko, R.R.; Feaver, M.; Bozkurt, B.T.; Lowe, N.; Nguyen, B.; Huang, K.W.; Wang, A.; Carney, R.P. A critical systematic review of extracellular vesicle clinical trials. J. Extracell. Vesicles 2024, 13, e12510. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.-P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of the first phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef]
- Viaud, S.; Ploix, S.; Lapierre, V.; Théry, C.; Commere, P.-H.; Tramalloni, D.; Gorrichon, K.; Virault-Rocroy, P.; Tursz, T.; Lantz, O.; et al. Updated technology to produce highly immunogenic dendritic cell-derived exosomes of clinical grade: A critical role of interferon-γ. J. Immunother. 2011, 34, 65–75. [Google Scholar] [CrossRef]
- Motawi, T.M.K.; Sabry, D.; Maurice, N.W.; Rizk, S.M. Role of mesenchymal stem cells exosomes derived microRNAs; miR-136, miR-494, and miR-495 in pre-eclampsia diagnosis and evaluation. Arch. Biochem. Biophys. 2018, 659, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.K.; Vashukova, E.S.; Illarionov, R.A.; Maltseva, A.R.; Pachuliia, O.V.; Postnikova, T.B.; Glotov, A.S. Extracellular Vesicles as Biomarkers of Pregnancy Complications. Int. J. Mol. Sci. 2024, 25, 11944. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Park, K.; Janne, P.A.; Kim, D.W.; Han, J.Y.; Wu, M.F.; Lee, J.S.; Kang, J.H.; Lee, D.H.; Cho, B.C.; Yu, C.J.; et al. Olmutinib in T790M-positive non-small cell lung cancer after failure of first-line epidermal growth factor receptor-tyrosine kinase inhibitor therapy: A global, phase 2 study. Cancer 2021, 127, 1407–1416. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical utility of the exosome-based ExoDx Prostate(IntelliScore) EPI test in men presenting for initial Biopsy with a PSA 2–10 ng/mL. Prostate Cancer Prostatic Dis. 2020, 23, 607–614. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McKiernan, J.; Donovan, M.J.; Margolis, E.; Partin, A.; Carter, B.; Brown, G.; Torkler, P.; Noerholm, M.; Skog, J.; Shore, N.; et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-Grade Prostate Cancer in Patients with Prostate-Specific Antigen 2–10 ng/mL at Initial Biopsy. Eur. Urol. 2018, 74, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Eitan, E.; Green, J.; Bodogai, M.; Mode, N.A.; Bæk, R.; Jørgensen, M.M.; Freeman, D.W.; Witwer, K.W.; Zonderman, A.B.; Biragyn, A.; et al. Age-Related Changes in Plasma Extracellular Vesicle Characteristics and Internalization by Leukocytes. Sci. Rep. 2017, 7, 1342. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, R.; Greening, D.W.; Zhu, H.J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Invest. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yekula, A.; Muralidharan, K.; Kang, K.M.; Wang, L.; Balaj, L.; Carter, B.S. From laboratory to clinic: Translation of extracellular vesicle-based cancer biomarkers. Methods 2020, 177, 58–66. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mattes, W.B.; Goodsaid, F. Regulatory landscapes for biomarkers and diagnostic tests: Qualification, approval, and role in clinical practice. Exp. Biol. Med. 2018, 243, 256–261. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Busatto, S.; Pham, A.; Suh, A.; Shapiro, S.; Wolfram, J. Organotropic drug delivery: Synthetic nanoparticles and extracellular vesicles. Biomed. Microdevices 2019, 21, 46. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sharma, S.; Masud, M.K.; Kaneti, Y.V.; Rewatkar, P.; Koradia, A.; Hossain, M.S.A.; Yamauchi, Y.; Popat, A.; Salomon, C. Extracellular Vesicle Nanoarchitectonics for Novel Drug Delivery Applications. Small 2021, 17, 2102220. [Google Scholar] [CrossRef] [PubMed]
- Busatto, S.; Walker, S.A.; Grayson, W.; Pham, A.; Tian, M.; Nesto, N.; Barklund, J.; Wolfram, J. Lipoprotein-based drug delivery. Adv. Drug Deliv. Rev. 2020, 159, 377–390. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, C.K.; Tsai, T.H.; Lee, C.H. Regulation of exosomes as biologic medicines: Regulatory challenges faced in exosome development and manufacturing processes. Clin. Transl. Sci. 2024, 17, e13904. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Calza, F.; Ferretti, M.; Panetti, E.; Parmentola, A. Moving drug discoveries beyond the valley of death: The role of innovation ecosystems. Eur. J. Innov. Manag. 2021, 24, 1184–1209. [Google Scholar] [CrossRef]
- Choi, A.; Koch, M.; Wu, K.; Dixon, G.; Oestreicher, J.; Legault, H.; Stewart-Jones, G.B.E.; Colpitts, T.; Pajon, R.; Bennett, H.; et al. Serum Neutralizing Activity of mRNA-1273 against SARS-CoV-2 Variants. J. Virol. 2021, 95, e0131321. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Turner, J.S.; O’Halloran, J.A.; Kalaidina, E.; Kim, W.; Schmitz, A.J.; Zhou, J.Q.; Lei, T.; Thapa, M.; Chen, R.E.; Case, J.B.; et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 2021, 596, 109–113. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Del Real Mata, C.; Jeanne, O.; Jalali, M.; Lu, Y.; Mahshid, S. Nanostructured-Based Optical Readouts Interfaced with Machine Learning for Identification of Extracellular Vesicles. Adv. Healthc. Mater. 2023, 12, e2202123. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A. Lost in translation: The valley of death across preclinical and clinical divide—Identification of problems and overcoming obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
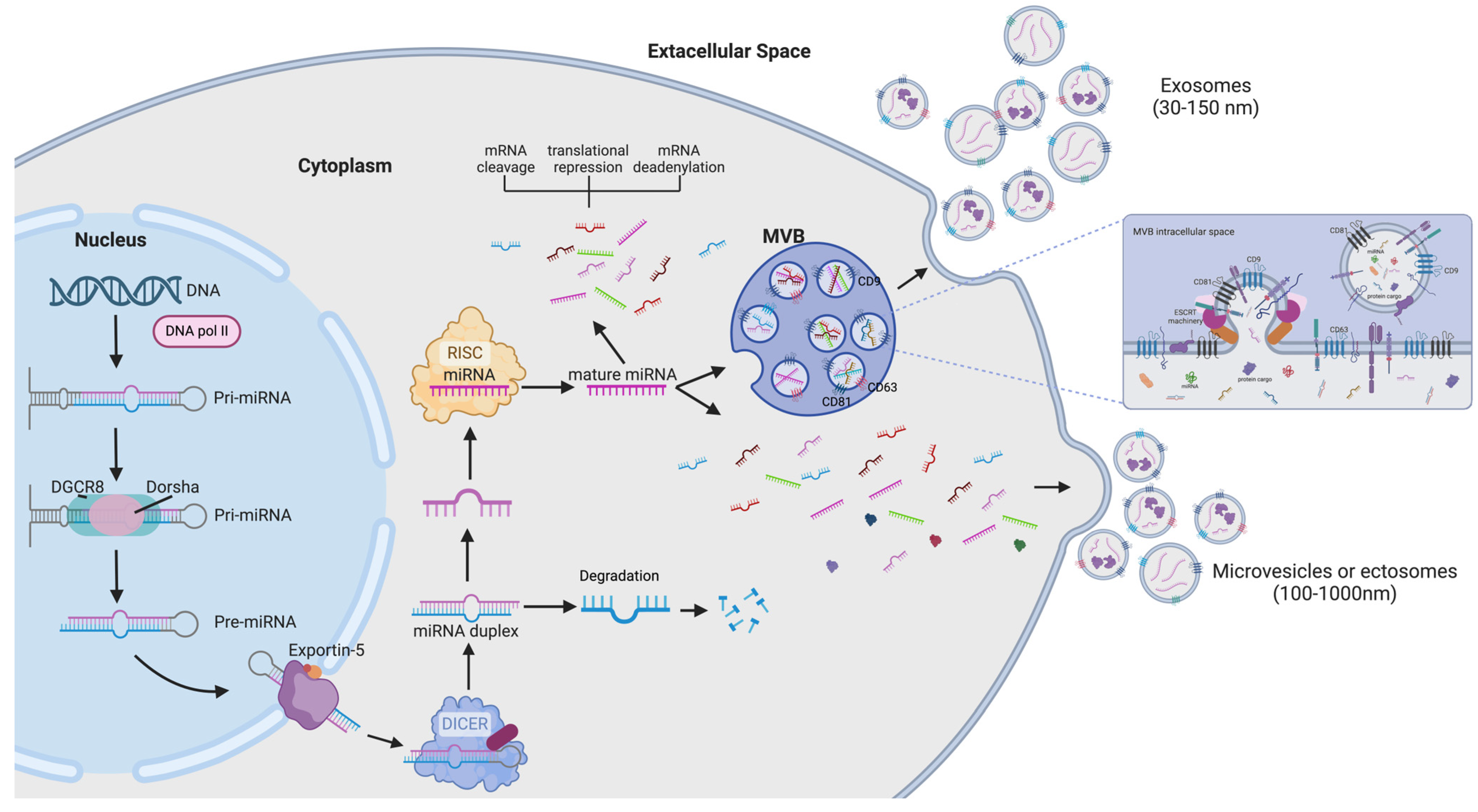
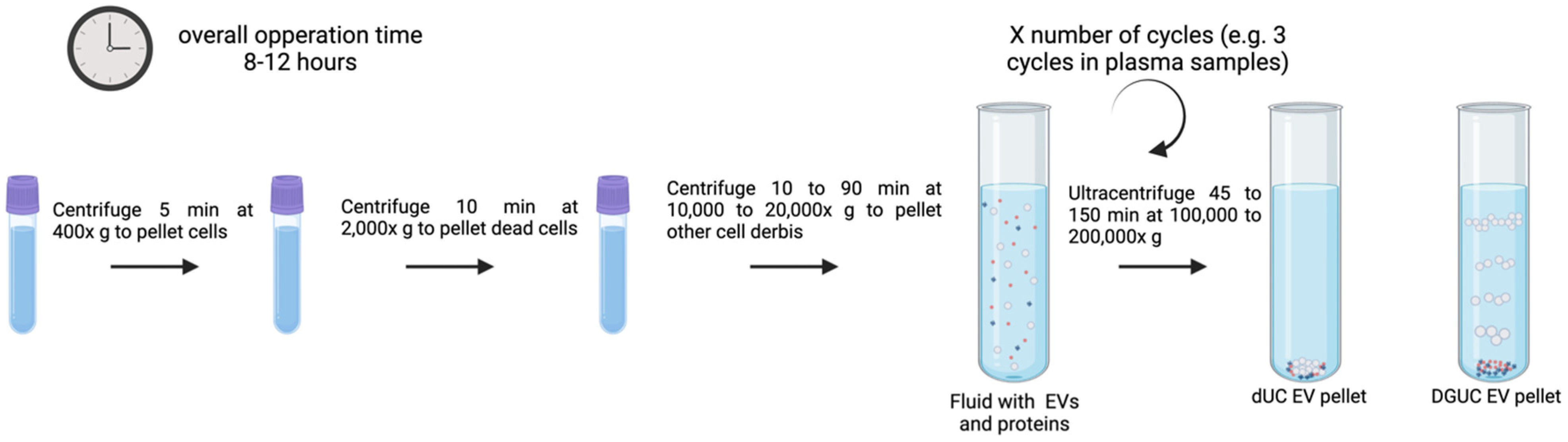
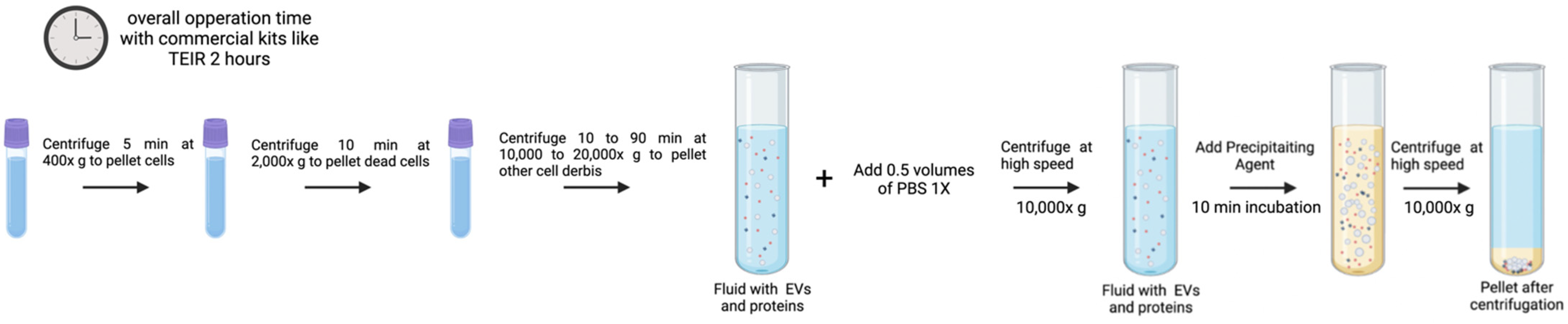


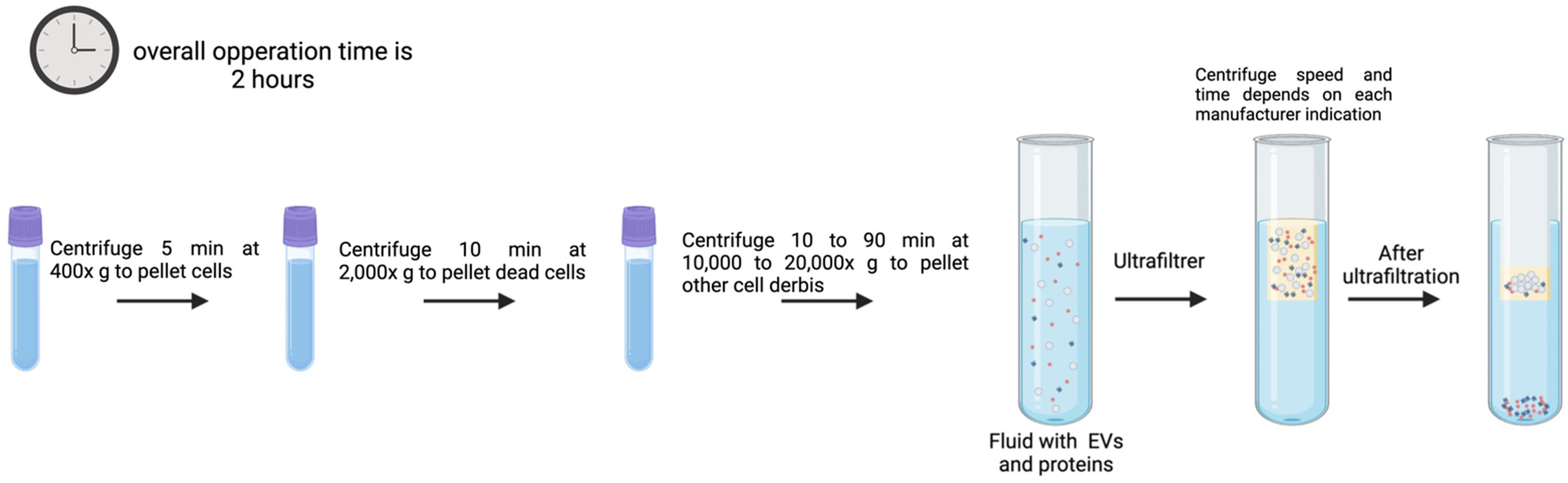
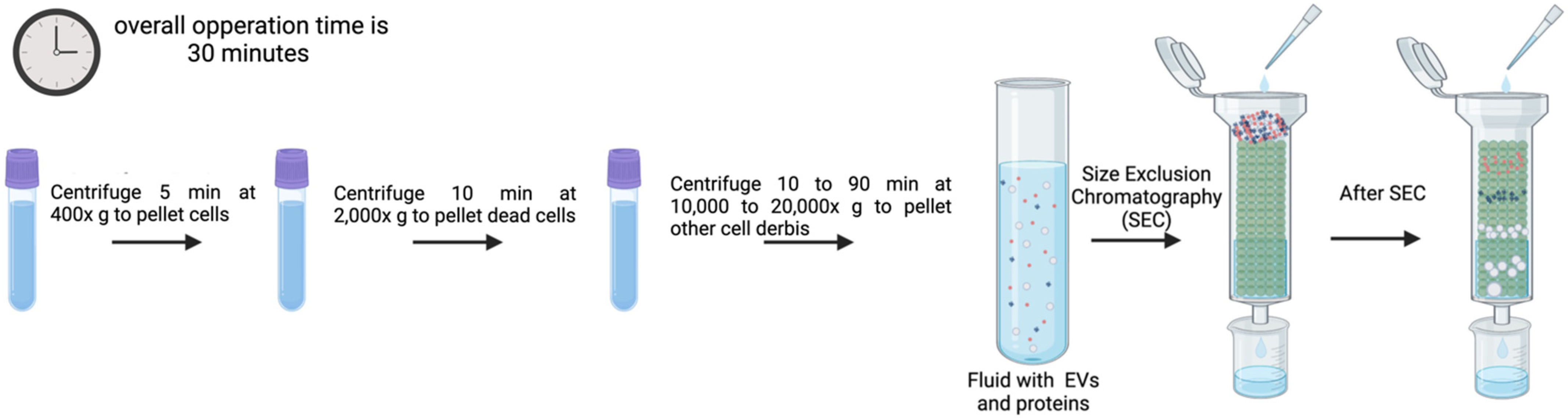
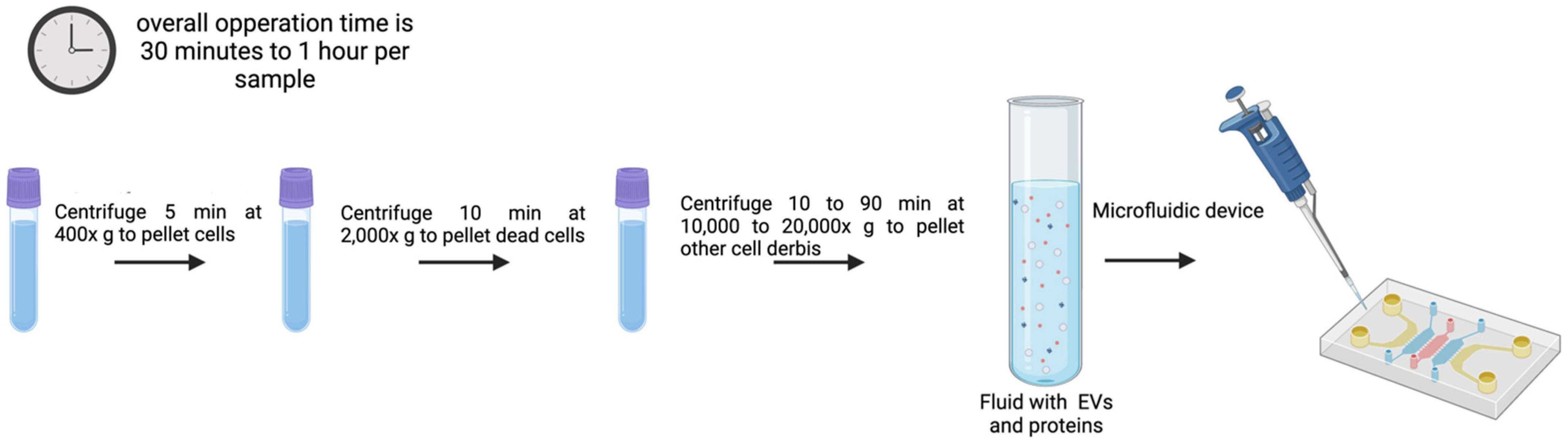
| Isolation Method | Advantages | Disadvantages | References |
|---|---|---|---|
| UC | Large sample amounts can be obtained. Easy to use. Low processing costs, capacity to handle large sample volumes, simultaneous separation of multiple EV samples, and no additional reagents are needed. DGUC: EV subtype isolation capacity. | Requirement of specialized equipment. EV structure may be disrupted. EV loss, fusion, distortion, and co-isolation of contaminants. Requires large sample volumes. Time-consuming. Dependent on the rotor, the temperature, and viscosity of the sample. Exosome aggregation can be induced. DGUC: Additional purification procedures are required to remove gradient solution in downstream applications. | [70,71,72,73,77] |
| Precipitation | Does not require specialized equipment, is quick, simple, affordable, and the volume required is low. EVs are not damaged. High isolation yield. | Poor EV purity, and protein contamination is generally high (pretreatment with proteases might help). | [82,83] |
| Immunoaffinity | High purity of isolated EVs by a rather simple process. | EV structure can be impacted by the non-neutral pH and non-physiological elution buffers. | [93,98] |
| EV sorting | Enables high-throughput analysis and categorization of EV based on biomarker expression. | High costs for modifying equipment and time-consuming. Detection limits in particle size. | [102,103] |
| UF | Fast, simple, and quick. It does not require special equipment. | It can damage EVs from shear stress, particle aggregation might compromise EV yields and consistency. | [67,97,106] |
| SEC | EVs maintain their integrity, important for biological activity assessment assays. High yields and low contamination. | Optimized SEC columns according to sample volume and type are required. | [113,114] |
| Microfluidics | Very high purity and recovery rate. EVs maintain their biological function. | Time-consuming | [131] |
| miR-15a and miR-16 | Chronic Lymphocytic Leukemia | qRT-PCR | [208] |
| miR-21 | Various Cancers (e.g., breast, lung, prostate) | qRT-PCR | [209] |
| miR-126 | Lung Cancer | Microarray Analysis | [210] |
| miR-122 | Hepatocellular Carcinoma | Northern Blot and qRT-PCR | [211] |
| miR-155 | Diffuse Large B-Cell Lymphoma | qRT-PCR | [212] |
| miR-21, miR-126, miR-146a | COVID-19 | qRT-PCR | [213] |
| miR-196b, miR-31, miR-891a, miR-34c, miR-653 | Lung Adenocarcinoma | Transcriptome Analysis | [214] |
| miR-21, miR-155 | Breast Cancer | Electrochemical Biosensors | [215] |
| miR-122, miR-192 | Hepatocellular Carcinoma | NGS | [216] |
| miR-29a, miR-181b | Alzheimer’s Disease | Surface-Enhanced Raman Scattering (SERS) Biosensors | [217] |
| Detection Method | Advantages | Disadvantages | Reference |
|---|---|---|---|
| qRT-PCR | High sensitivity and specificity. Quantitative and widely used. | Requires prior sequence knowledge. Limited detection of novel miRNAs. | [232] |
| Microarray Analysis | High-throughput detection of multiple miRNAs. Suitable for comparative expression profiling. | Lower sensitivity than qRT-PCR. Detects only known miRNAs. | [231] |
| NGS | Allows discovery of novel miRNAs. High sensitivity and dynamic range. | Expensive and requires complex bioinformatics. Long turnaround time. | [236] |
| ISH | Provides spatial distribution of miRNA expression. Single-cell resolution. | Less quantitative than qRT-PCR/NGS. Requires high-quality tissue samples. | [240,241] |
| NB | Confirms miRNA integrity and size. | Labor-intensive and requires large RNA amounts. Low sensitivity. | [245,248] |
| Biosensors | Rapid and real-time detection. Potential for portable diagnostics. | Requires careful design and optimization. May be affected by biological sample complexity. | [250,257,261,262] |
| ddPCR | Absolute quantification without a standard curve. High sensitivity, even for low-abundance miRNAs. Resistant to PCR inhibitors. | More expensive than qRT-PCR. Limited multiplexing capabilities. | [257,258] |
| NanoString | Direct and absolute quantification. High specificity due to sequence-specific probes. Multiplexing capability. Works well with low RNA input and degraded samples. High reproducibility and ease of use. | Lower sensitivity compared to qPCR for low-abundance miRNAs. Higher cost per sample compared to some qPCR-based methods. Requires specialized equipment (nCounter system). | [259,260] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ljungström, M.; Oltra, E. Methods for Extracellular Vesicle Isolation: Relevance for Encapsulated miRNAs in Disease Diagnosis and Treatment. Genes 2025, 16, 330. https://doi.org/10.3390/genes16030330
Ljungström M, Oltra E. Methods for Extracellular Vesicle Isolation: Relevance for Encapsulated miRNAs in Disease Diagnosis and Treatment. Genes. 2025; 16(3):330. https://doi.org/10.3390/genes16030330
Chicago/Turabian StyleLjungström, Maria, and Elisa Oltra. 2025. "Methods for Extracellular Vesicle Isolation: Relevance for Encapsulated miRNAs in Disease Diagnosis and Treatment" Genes 16, no. 3: 330. https://doi.org/10.3390/genes16030330
APA StyleLjungström, M., & Oltra, E. (2025). Methods for Extracellular Vesicle Isolation: Relevance for Encapsulated miRNAs in Disease Diagnosis and Treatment. Genes, 16(3), 330. https://doi.org/10.3390/genes16030330






