Transcriptome and Metabolome Analysis of Low-Pressure Regulation in Saussurea involucrata Leaves
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Changes in S. involucrata Leaves Under Different Air-Pressure Treatments
2.2. Transcriptome Analysis of the S. involucrata
2.2.1. Transcriptome Data-Quality Assessment of the S. involucrata and Identification of DEGs
2.2.2. Enrichment Analysis of DEGs
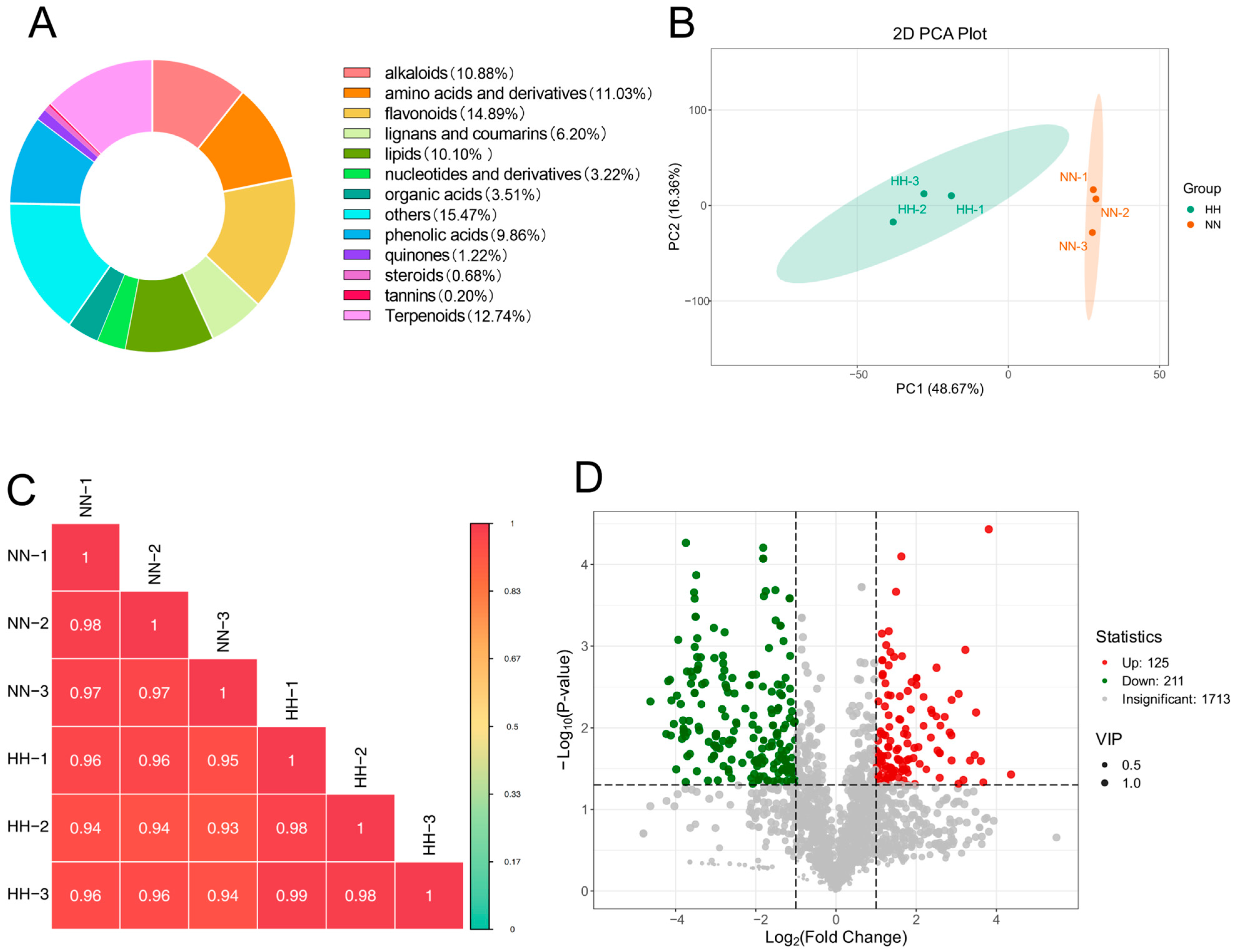
2.3. Metabolome Analysis of the S. involucrata
2.3.1. Detection and Screening of DEGs
2.3.2. KEGG Enrichment Analysis of DAMs
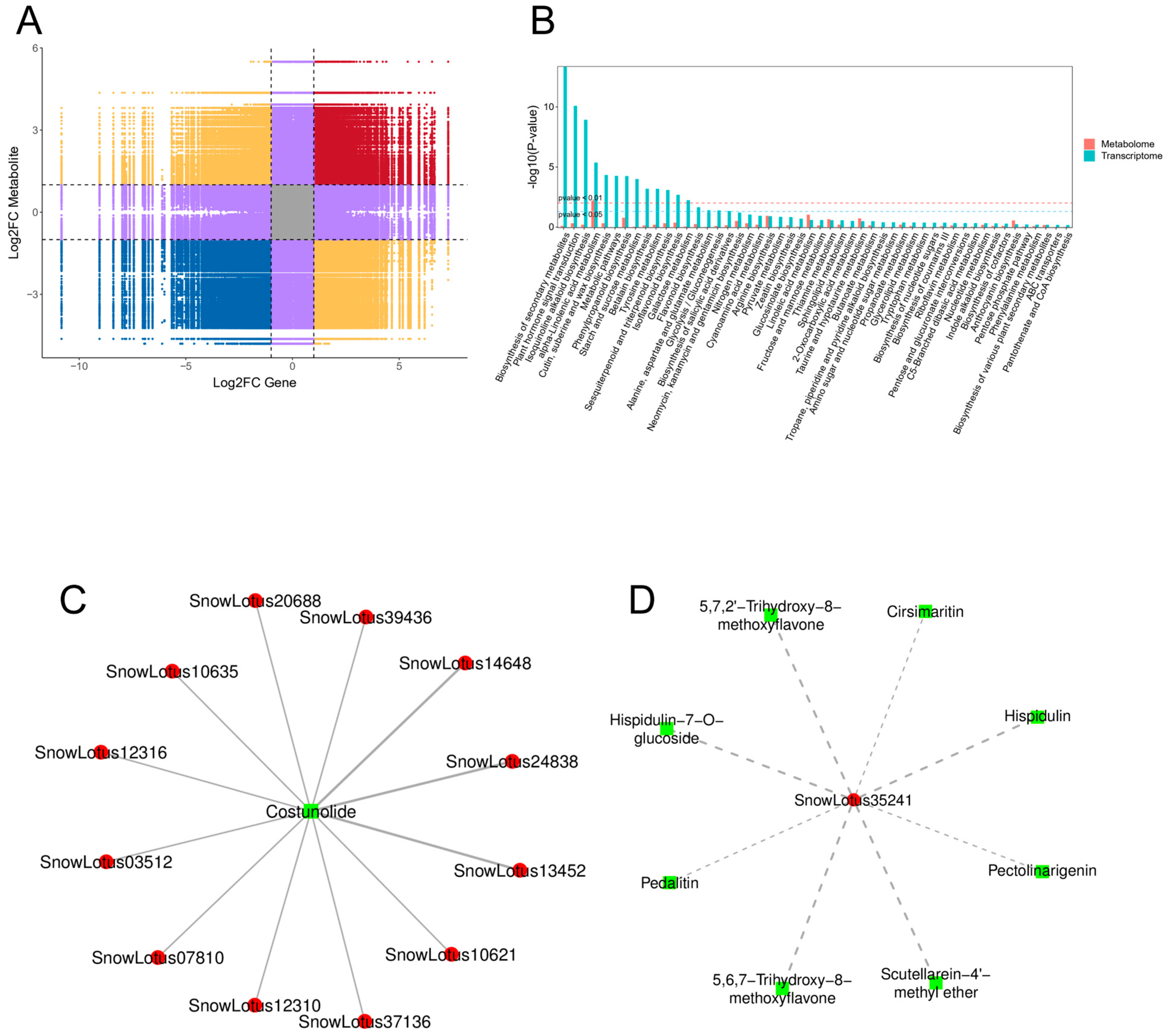
2.4. Combined Analysis of the DEGs and DAMs
2.4.1. DEG–DAM Correlation Analysis
2.4.2. Key Metabolic Pathways Influenced by Low Pressure
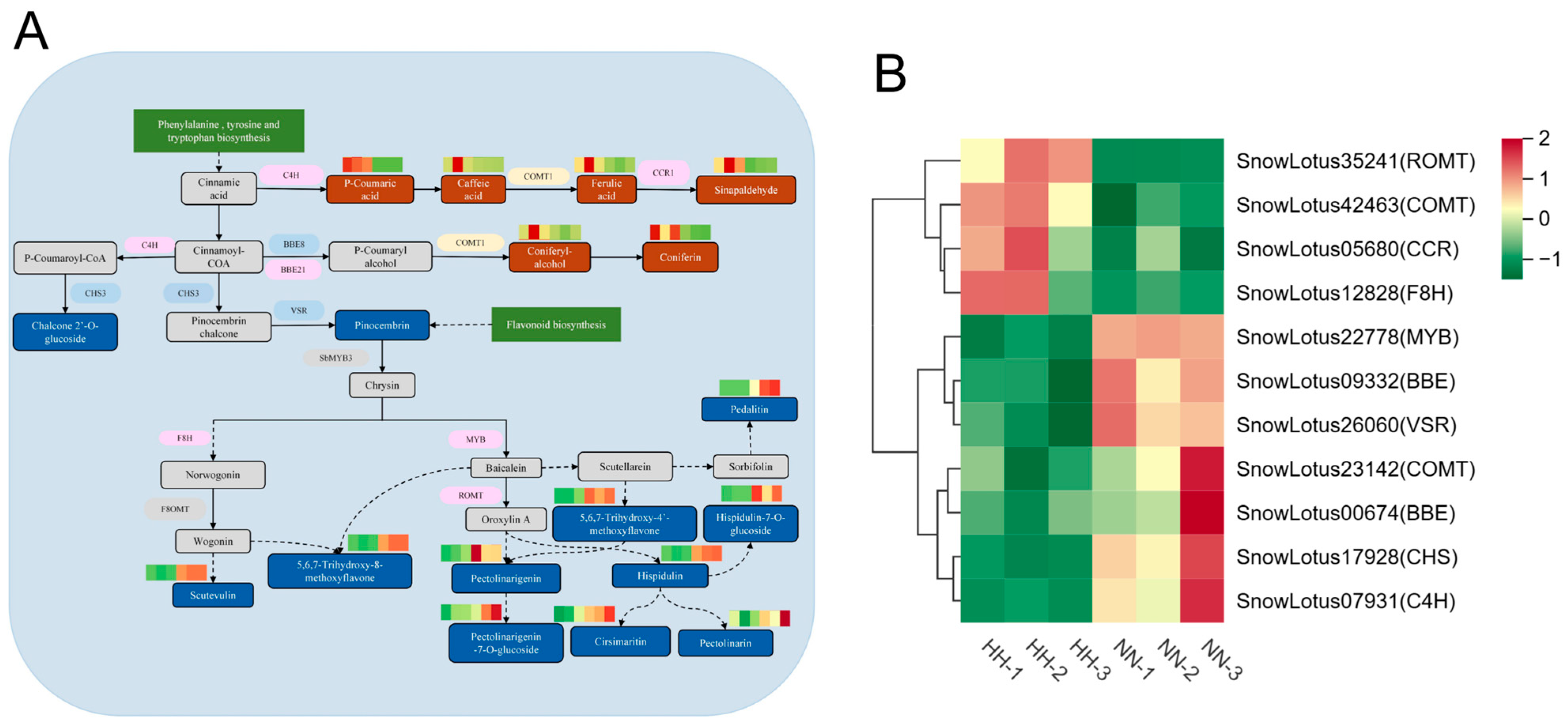
2.4.3. Role of Flavonoids and Phenylpropanoids in Hypoxia Adaptation
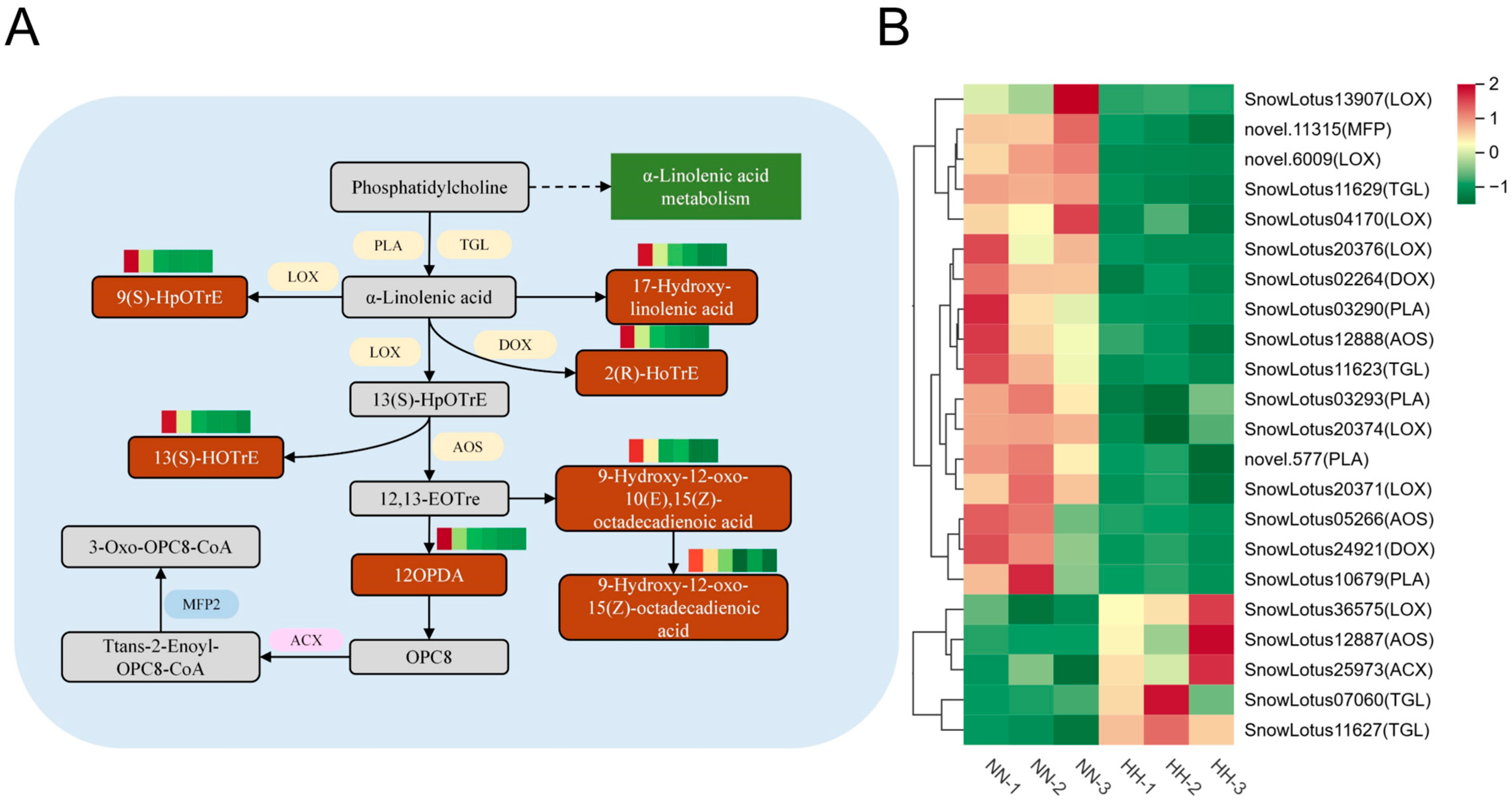
2.4.4. Lipid Metabolism and Stress Response Mechanisms
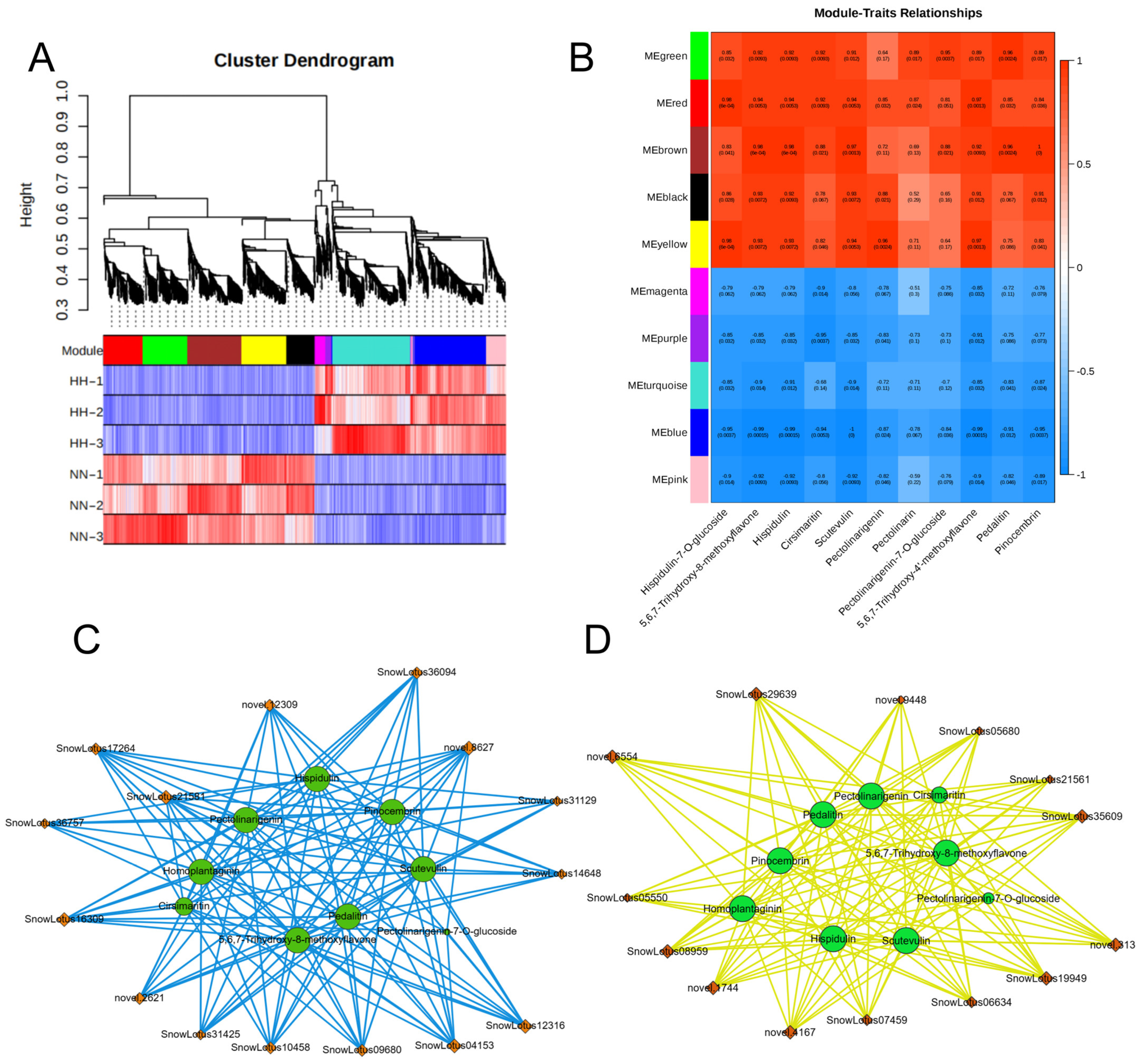
2.4.5. Identification of Hub Genes and Their Functional Significance
3. Discussion
3.1. A Multi-Omics Study of Hypobaric Hypoxia Adaptation in S. involucrata
3.2. Reprogramming of Flavonoid Metabolism for Hypobaric Hypoxia Acclimation in S. involucrata
3.3. JA-Mediated α-Linolenic Acid Antioxidant Defense in S. involucrata Under Hypobaric Hypoxia
3.4. Coordinated Regulation of Hypobaric Hypoxia Acclimation in S. involucrata
4. Materials and Methods
4.1. Treatment of S. involucrata
4.2. RNA Extraction and cDNA Reversal
4.3. Sequence Data Processing and Analysis of DEGs
4.4. Metabolite Extraction and DAM Analysis
4.5. Combined Transcriptome and Metabolome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gale, J. Availability of carbon dioxide for photosynthesis at high altitudes: Theoretical considerations. Ecology 1972, 53, 494–497. [Google Scholar] [CrossRef]
- He, C.; Davies, F.T., Jr.; Lacey, R.E. Ethylene reduces gas exchange and growth of lettuce plants under hypobaric and normal atmospheric conditions. Physiol. Plant. 2009, 135, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.-L.; Schuerger, A.C.; Popp, M.P.; Richards, J.T.; Manak, M.S.; Ferl, R.J. Hypobaric Biology: Arabidopsis Gene Expression at Low Atmospheric Pressure1 [w]. Plant Physiol. 2004, 134, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [CrossRef] [PubMed]
- Branco-Price, C.; Kawaguchi, R.; Ferreira, R.B.; Bailey-Serres, J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 2005, 96, 647–660. [Google Scholar] [CrossRef]
- Klok, E.J.; Wilson, I.W.; Wilson, D.; Chapman, S.C.; Ewing, R.M.; Somerville, S.C.; Peacock, W.J.; Dolferus, R.; Dennis, E.S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 2002, 14, 2481–2494. [Google Scholar] [CrossRef]
- Liu, F.; VanToai, T.; Moy, L.P.; Bock, G.; Linford, L.D.; Quackenbush, J. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 2005, 137, 1115–1129. [Google Scholar] [CrossRef]
- Rathore, N.; Kumar, P.; Mehta, N.; Swarnkar, M.K.; Shankar, R.; Chawla, A. Time-series RNA-Seq transcriptome profiling reveals novel insights about cold acclimation and de-acclimation processes in an evergreen shrub of high altitude. Sci. Rep. 2022, 12, 15553. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, Y.; Landis, J.B.; Shen, J.; Zhang, H.; Kuang, T.; Sun, W.; Sun, J.; Tiamiyu, B.B.; Deng, T. Transcriptomes of Saussurea (Asteraceae) provide insights into high-altitude adaptation. Plants 2021, 10, 1715. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Ma, L.; Sun, X.; Kong, X.; Galvan, J.V.; Li, X.; Yang, S.; Yang, Y.; Yang, Y.; Hu, X. Physiological, biochemical and proteomics analysis reveals the adaptation strategies of the alpine plant Potentilla saundersiana at altitude gradient of the Northwestern Tibetan Plateau. J. Proteom. 2015, 112, 63–82. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yang, L.; Zhao, J.; Wei, J.; Kong, X.; Wang, C.; Zhang, X.; Yang, Y.; Hu, X. Comparative proteomic analysis reveals the role of hydrogen sulfide in the adaptation of the alpine plant Lamiophlomis rotata to altitude gradient in the Northern Tibetan Plateau. Planta 2015, 241, 887–906. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, Y.; Lyu, B.; Chen, J.; Chen, G. Component Identification of Phenolic Acids in Cell Suspension Cultures of Saussurea involucrata and Its Mechanism of Anti-Hepatoma Revealed by TMT Quantitative Proteomics. Foods 2021, 10, 2466. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-C.; Kao, J.-Y.; Kuo, D.-H.; Liao, C.-F.; Huang, C.-H.; Fan, L.-L.; Way, T.-D. Antifatigue and antioxidant activity of alcoholic extract from Saussurea involucrata. J. Tradit. Complement. Med. 2011, 1, 64–68. [Google Scholar] [CrossRef]
- Chen, J.; Wu, X.; Yu, R. Unraveling the Therapeutic Mechanism of Saussurea involucrata against Rheumatoid Arthritis: A Network Pharmacology and Molecular Modeling-Based Investigation. Nutrients 2023, 15, 4294. [Google Scholar] [CrossRef]
- Wang, D.; Jin, Y.; Yang, M.; Xue, Y.; Zhang, X.; Guo, Y.; Li, X.; Ma, K. Cardioprotective effect of Saussurea involucrata injection against Doxorubicin-induced cardiotoxicity by network pharmacology analysis and experimental verification. Acta Biochim. Biophys. Sin. 2024. [Google Scholar] [CrossRef]
- Wang, X.; Chu, L.; Liu, C.; Wei, R.; Xue, X.; Xu, Y.; Wu, M.; Miao, Q. Therapeutic effects of Saussurea involucrata injection against severe acute pancreatitis-induced brain injury in rats. Biomed. Pharmacother. 2018, 100, 564–574. [Google Scholar] [CrossRef]
- Chik, W.-I.; Zhu, L.; Fan, L.-L.; Yi, T.; Zhu, G.-Y.; Gou, X.-J.; Tang, Y.-N.; Xu, J.; Yeung, W.-P.; Zhao, Z.-Z. Saussurea involucrata: A review of the botany, phytochemistry and ethnopharmacology of a rare traditional herbal medicine. J. Ethnopharmacol. 2015, 172, 44–60. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.-J. New Saussurea (Asteraceae) species from Bogeda Mountain, eastern Tianshan, China, and inference of its evolutionary history and medical usage. PLoS ONE 2018, 13, e0199416. [Google Scholar] [CrossRef]
- Hu, L.; Lu, T.; Wang, X.; Wang, J.; Shi, W. Conservation Priorities and Demographic History of Saussurea involucrata in the Tianshan Mountains and Altai Mountains. Life 2023, 13, 2209. [Google Scholar] [CrossRef]
- Nyamgerel, N.; Baasanmunkh, S.; Oyuntsetseg, B.; Tsegmed, Z.; Bayarmaa, G.-A.; Lazkov, G.; Pyak, E.; Gil, H.-Y.; Park, I.; Choi, H.J. Comparative plastome analysis and taxonomic classification of snow lotus species (Saussurea, Asteraceae) in Central Asia and Southern Siberia. Funct. Integr. Genom. 2024, 24, 42. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Fu, Y.; Liu, B.; Zhang, Y.; Wang, A.; Li, Y.; Zhu, J. SiFBA5, a cold-responsive factor from Saussurea involucrata promotes cold resilience and biomass increase in transgenic tomato plants under cold stress. BMC Plant Biol. 2021, 21, 75. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, L.; Wang, X.; Zhang, M.; Xi, Y.; Wang, A.; Zhu, J. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, W.; Huang, G.; Liu, B.; Wang, A.; Zhu, J.; Guo, X. The SikCuZnSOD3 gene improves abiotic stress resistance in transgenic cotton. Mol. Breed. 2021, 41, 26. [Google Scholar] [CrossRef]
- Kreuzwieser, J.; Hauberg, J.; Howell, K.A.; Carroll, A.; Rennenberg, H.; Millar, A.H.; Whelan, J. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009, 149, 461–473. [Google Scholar] [CrossRef]
- Safavi-Rizi, V.; Herde, M.; Stöhr, C. RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root. Sci. Rep. 2020, 10, 1692. [Google Scholar]
- Sanclemente, M.-A.; Ma, F.; Liu, P.; Della Porta, A.; Singh, J.; Wu, S.; Colquhoun, T.; Johnson, T.; Guan, J.-C.; Koch, K.E. Sugar modulation of anaerobic-response networks in maize root tips. Plant Physiol. 2021, 185, 295–317. [Google Scholar] [CrossRef]
- Grandbastien, M.-A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998, 3, 181–187. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, X.; Chen, X.; Wang, B.; Dong, M.; Chen, L.; Yang, Z.; Li, Y.; Sun, H. Integrated Untargeted Metabolome, Full-Length Sequencing and Transcriptome Analyses Reveal the Mechanism of Flavonoid Biosynthesis in Blueberry (Vaccinium spp.) Fruit. Int. J. Mol. Sci. 2024, 25, 4137. [Google Scholar] [CrossRef]
- Upchurch, R.G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 2008, 30, 967–977. [Google Scholar] [CrossRef]
- Xu, J.; Chen, W.-J.; Wang, Z.; Xin, M.-Y.; Gao, S.-H.; Liu, W.-J.; Wang, K.-K.; Ma, J.-W.; Yan, X.-Z.; Ren, Y.-M. Profiles of transcriptome and metabolic pathways after hypobaric hypoxia exposure. Proteome Sci. 2022, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zhou, Z.; Mei, Z. Integrative analysis of metabolome and transcriptome identifies potential genes involved in the flavonoid biosynthesis in Entada phaseoloides stem. Front. Plant Sci. 2022, 13, 792674. [Google Scholar] [CrossRef]
- Xu, F.; Ning, Y.; Zhang, W.; Liao, Y.; Li, L.; Cheng, H.; Cheng, S. An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba. Funct. Integr. Genom. 2014, 14, 177–189. [Google Scholar] [CrossRef]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Luo, F.-L.; Thiele, B.; Janzik, I.; Zeng, B.; Schurr, U.; Matsubara, S. De-submergence responses of antioxidative defense systems in two wetland plants having escape and quiescence strategies. J. Plant Physiol. 2012, 169, 1680–1689. [Google Scholar] [CrossRef]
- Yuan, L.-B.; Dai, Y.-S.; Xie, L.-J.; Yu, L.-J.; Zhou, Y.; Lai, Y.-X.; Yang, Y.-C.; Xu, L.; Chen, Q.-F.; Xiao, S. Jasmonate regulates plant responses to postsubmergence reoxygenation through transcriptional activation of antioxidant synthesis. Plant Physiol. 2017, 173, 1864–1880. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, R.; Xiang, C.; Zhang, R.; Wang, Q.; Wang, T.; Li, X.; Lu, X.; Gao, S.; Liu, Z. Transcriptomic and physiological analysis reveal that α-linolenic acid biosynthesis responds to early chilling tolerance in pumpkin rootstock varieties. Front. Plant Sci. 2021, 12, 669565. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Handa, A.K.; Mattoo, A.K. Transcript abundance patterns of 9-and 13-lipoxygenase subfamily gene members in response to abiotic stresses (heat, cold, drought or salt) in tomato (Solanum lycopersicum L.) highlights member-specific dynamics relevant to each stress. Genes 2019, 10, 683. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yamamoto, A.; Nakamura, T.; Nouri, M.-Z.; Nanjo, Y.; Nishizawa, K.; Furukawa, K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteome Res. 2011, 10, 3993–4004. [Google Scholar] [CrossRef] [PubMed]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Callaham, J.B.; Reyes, M.; Stasiak, M.; Riva, A.; Zupanska, A.K.; Dixon, M.A.; Paul, A.-L.; Ferl, R.J. Dissecting low atmospheric pressure stress: Transcriptome responses to the components of hypobaria in Arabidopsis. Front. Plant Sci. 2017, 8, 528. [Google Scholar] [CrossRef]
- Mir, R.R.; Reynolds, M.; Pinto, F.; Khan, M.A.; Bhat, M.A. High-throughput phenotyping for crop improvement in the genomics era. Plant Sci. 2019, 282, 60–72. [Google Scholar] [CrossRef]
- Husain, T.; Fatima, A.; Suhel, M.; Singh, S.; Sharma, A.; Prasad, S.M.; Singh, V.P. A brief appraisal of ethylene signaling under abiotic stress in plants. Plant Signal. Behav. 2020, 15, 1782051. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Hsu, F.-C.; Li, J.-P.; Wang, N.-N.; Shih, M.-C. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011, 156, 202–212. [Google Scholar] [CrossRef]
- Imani Asl, E.; Soorni, A.; Mehrabi, R. Genome-wide characterization, functional analysis, and expression profiling of the Aux/IAA gene family in spinach. BMC Genom. 2024, 25, 567. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Gustavsen, J.A.; Pai, S.; Isserlin, R.; Demchak, B.; Pico, A.R. RCy3: Network biology using Cytoscape from within R. F1000Research 2019, 8, 1774. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quan, X.; Fan, F.; Cao, H.; Tang, N.; Xu, C.; Wang, C. Transcriptome and Metabolome Analysis of Low-Pressure Regulation in Saussurea involucrata Leaves. Genes 2025, 16, 328. https://doi.org/10.3390/genes16030328
Quan X, Fan F, Cao H, Tang N, Xu C, Wang C. Transcriptome and Metabolome Analysis of Low-Pressure Regulation in Saussurea involucrata Leaves. Genes. 2025; 16(3):328. https://doi.org/10.3390/genes16030328
Chicago/Turabian StyleQuan, Xinyu, Fenggui Fan, Hanbo Cao, Na Tang, Changgen Xu, and Changhe Wang. 2025. "Transcriptome and Metabolome Analysis of Low-Pressure Regulation in Saussurea involucrata Leaves" Genes 16, no. 3: 328. https://doi.org/10.3390/genes16030328
APA StyleQuan, X., Fan, F., Cao, H., Tang, N., Xu, C., & Wang, C. (2025). Transcriptome and Metabolome Analysis of Low-Pressure Regulation in Saussurea involucrata Leaves. Genes, 16(3), 328. https://doi.org/10.3390/genes16030328





