Animal Species and Identity Testing: Developments, Challenges, and Applications to Non-Human Forensics
Abstract
1. Introduction
2. Challenges in Wildlife Genetic Identity Testing
- (a)
- What is the goal, i.e., resolution needed for the specific case?
- (b)
- Are the samples provided sufficient for analysis?
- (c)
- Are the data probative?
- (d)
- Does the sample contain biological material from an endangered species?
- (e)
- Is damage to the artifact during sampling a constraint?
- (f)
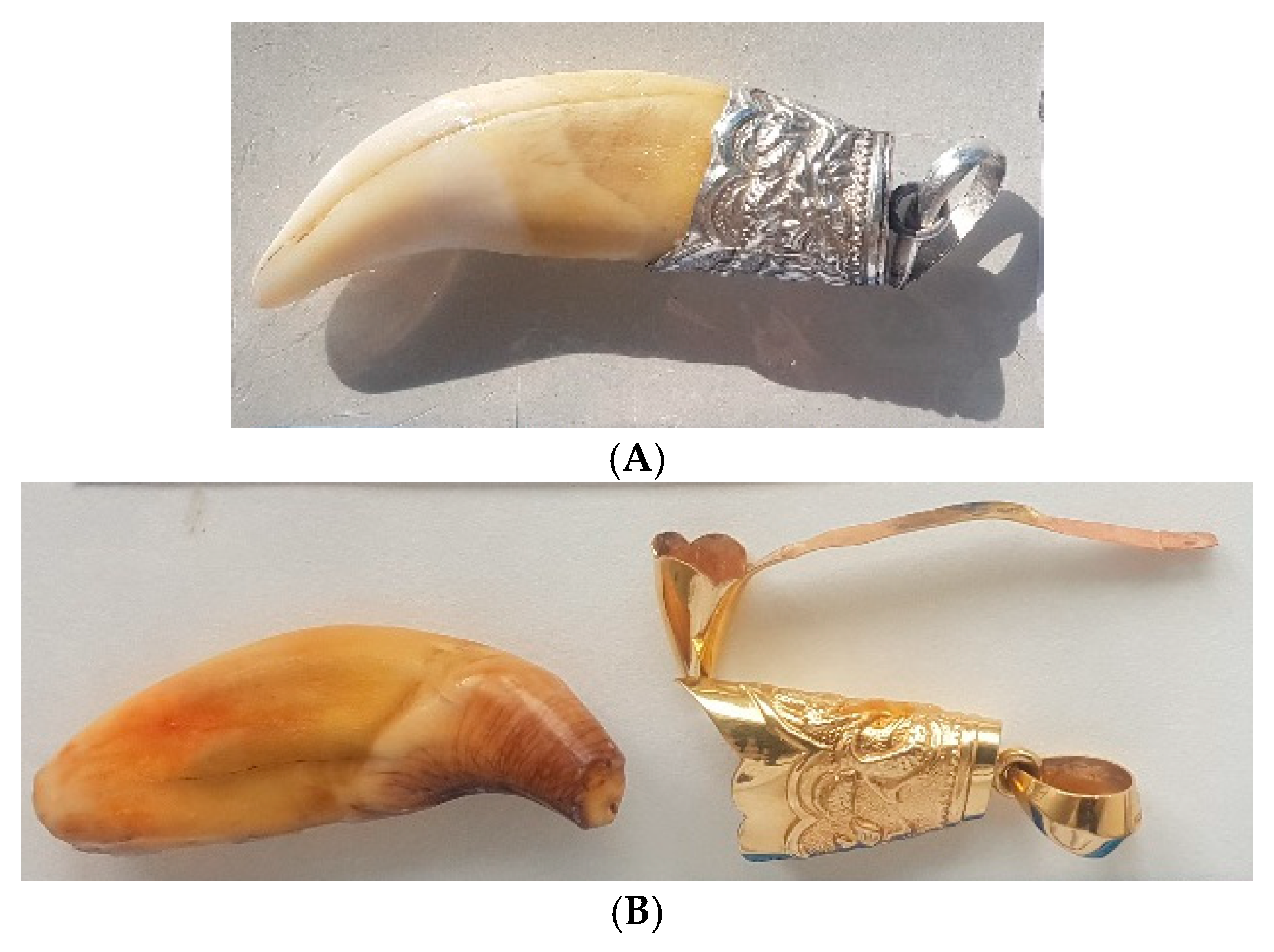
- Is the artifact made from only one animal?
- (g)
- Was the material chemically treated?
- (h)
- What extraction procedure(s) will be appropriate?
- (i)
- Which genetic marker(s) should be tested?
- (j)
- How should the data be interpreted?
- (k)
- Are there specific population genetics aspects that should be considered?
- (l)
- Does the background create noise?
2.1. Sampling
2.2. Environmental DNA
2.3. Post-Sampling DNA Stability
2.4. Human Contamination of Non-Human Samples
2.5. Analytical Methods
2.6. DNA Extraction and Quantitation
2.7. Primers for DNA Barcoding
2.8. DNA-Based Barcoding Methods
2.9. Nuclear Inserts of Mitochondrial Genome (NUMTs)
2.10. Hybrids
2.11. Individual Identification in Non-Human Forensic DNA Testing
2.12. ISO Standards
2.13. Legal and Ethical/Conduct Hurdles
3. Discussion
- Improve Sampling Procedures: For composite artifacts like taxidermy, sub-sample an item (bearing in mind constraints on damaging certain artifacts) to reduce the risk of false-positive or false-negative results. Use sampling kits containing reagents that stabilize nucleic acids and inactivate infectious agents (when appropriate), which are crucial for field-collected samples without cold-chain storage. Take special care to avoid contamination with human DNA, particularly when collection and/or sampling is performed by non-specialized personnel.
- Advance Laboratory Protocols: Develop and validate DNA extraction and inhibitor removal protocols specifically for non-human samples (such as those undertaken in microbial forensics applications), as human forensic methods may be insufficient for the task. Employ, when feasible, qPCR with an internal amplification control to accurately measure the target DNA and determine the impact of inhibitors in a sample. Produce robust enrichment kits that may be able to work in the presence of inhibitors at least to some degree. Acknowledge the limitations of mtDNA-based typing. Use, when possible, species-specific nuclear markers to confirm or exclude the presence of hybrids and sample mixtures. Embrace high-throughput sequencing methods to improve detection and sensitivity over current or traditional approaches. Acknowledge and explicitly state limitations in any statistical calculations and inferences.
- Enhance Standards and Databases: Develop and validate sampling and diagnostic kits specifically for non-human genetic identification, as current ISO standards, like ISO 18385, do not cover non-human DNA contaminants. Adhere to standards and recommendations from bodies like SWFS and ENFSI-APST, which provide guidelines for the field. If the restriction of shipping CITES-protected samples cannot be overcome, then pursue collaborative studies and/or use mimicked DNA. Alternatively, develop memoranda of understanding with in-country zoos or museums to obtain a number, albeit limited, of samples. Recognize that online genetic databases like GenBank and BOLD have varying levels of completeness, overlap with entries, and can contain errors, which should be considered during the development and validation of assays and during analysis. Genetic testing and database searches also serve as deterrents for potential offenders who may know they can be easily linked to crimes.
- Adopt a Multidisciplinary Approach: The spectrum of relevant samples in wildlife crime cases is vast. Additional methods, such as profiling of volatilomes [201], mass spectrometry [202], hair and feather morphology [203,204], osteology [205,206], radiocarbon dating [207], and the analysis of stable isotopes [208,209,210], to name a few, may provide faster answers than DNA analysis, or at least provide additional support for findings.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The International Consortium on Combating Wildlife Crime. Available online: https://cites.org/eng/prog/iccwc (accessed on 1 September 2025).
- Parties to CITES. Available online: https://cites.org/eng/disc/parties/index.php (accessed on 1 September 2025).
- Budowle, B.; Garofano, P.; Hellman, A.; Ketchum, M.; Kanthaswamy, S.; Parson, W.; Van Haeringen, W.; Fain, S.; Broad, T. Recommendations for animal DNA forensic and identity testing. Int. J. Leg. Med. 2005, 119, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Linacre, A.; Gusmão, L.; Hecht, W.; Hellmann, A.; Mayr, W.; Parson, W.; Prinz, M.; Schneider, P.; Morling, N. ISFG: Recommendations regarding the use of non-human (animal) DNA in forensic genetic investigations. Forensic Sci. Int. Genet. 2011, 5, 501–505. [Google Scholar] [CrossRef]
- Kanthaswamy, S. Wildlife forensic genetics—Biological evidence, DNA markers, analytical approaches, and challenges. Anim. Genet. 2024, 55, 177–192. [Google Scholar] [CrossRef]
- Ruedi, M.; Manzinalli, J.; Dietrich, A.; Vinciguerra, L. Shortcomings of DNA barcodes: A perspective from the mammal fauna of Switzerland. Hystrix Ital. J. Mammal. 2023, 34, 54–61. [Google Scholar]
- De Bruyn, M.; Dalton, D.L.; Harper, C.K.; Sethusa, M.T. A septennium review of wildlife forensic DNA analysis in South Africa. Forensic Sci. Int. Genet. 2025, 80, 103339. [Google Scholar] [CrossRef]
- Michel, A.L.; Van Heerden, H.; Prasse, D.; Rutten, V.P.; Al Dahouk, S.; Crossley, B. Pathogen detection and disease diagnosis in wildlife: Challenges and opportunities. Rev. Sci. Tech. l’OIE 2021, 40, 105–118. [Google Scholar] [CrossRef]
- Alacs, E.A.; Georges, A.; FitzSimmons, N.N.; Robertson, J. DNA detective: A review of molecular approaches to wildlife forensics. Forensic Sci. Med. Pathol. 2010, 6, 180–194. [Google Scholar] [CrossRef]
- Moore, M.K.; Frazier, K. Humans are animals, too: Critical commonalities and differences between human and wildlife forensic genetics. J. Forensic Sci. 2019, 64, 1603–1621. [Google Scholar] [CrossRef]
- Smart, U.; Cihlar, J.C.; Budowle, B. International Wildlife Trafficking: A perspective on the challenges and potential forensic genetics solutions. Forensic Sci. Int. Genet. 2021, 54, 102551. [Google Scholar] [CrossRef]
- Johnson, R.N.; Wilson-Wilde, L.; Linacre, A. Current and future directions of DNA in wildlife forensic science. Forensic Sci. Int. Genet. 2014, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.R.N.; Pinto, L.C.L.; Barboza, R.R.D.; Souto, W.M.S.; Oliveira, R.E.M.C.C.; Vieira, W.L.S. A global overview of carnivores used in traditional medicines. In Animals in Traditional Folk Medicine: Implications for Conservation; Springer: Berlin/Heidelberg, Germany, 2012; pp. 171–206. [Google Scholar]
- Byard, R.W. Traditional medicines and species extinction: Another side to forensic wildlife investigation. Forensic Sci. Med. Pathol. 2016, 12, 125–127. [Google Scholar] [CrossRef]
- Sharma, S.; Thokchom, R. A review on endangered medicinal plants of India and their conservation. J. Crop Weed 2014, 10, 205–218. [Google Scholar]
- Su, L.; Pan, Y. Ethical Considerations in the Use of Endangered Species for Traditional Chinese Medicine Practices: A Case Study of the Pangolin. J. Res. Soc. Sci. Humanit. 2024, 3, 24–30. [Google Scholar] [CrossRef]
- Shao, M.-L.; Newman, C.; Buesching, C.D.; Macdonald, D.W.; Zhou, Z.-M. Understanding wildlife crime in China: Socio-demographic profiling and motivation of offenders. PLoS ONE 2021, 16, e0246081. [Google Scholar] [CrossRef] [PubMed]
- Lennklo, D. Anthropomorphic Animals in Commercials: Why Fake Animals Tell Good Stories. Master’s Thesis, Lund University, Lund, Sweden, 2010. [Google Scholar]
- Hume, S. Could nature turn a hare into a jackalope? J. Interdiscip. Sci. Top. 2022, 9. Available online: https://journals.le.ac.uk/index.php/jist/article/view/4045 (accessed on 10 December 2025).
- Magdeburg Unicorn. Available online: https://www.naturkundemuseum-magdeburg.de/en/dauerausstellung/einhorn-skelett/ (accessed on 18 August 2025).
- Ariew, R. Leibniz on the Unicorn and various other curiosities. Early Sci. Med. 1998, 3, 267–288. [Google Scholar] [CrossRef]
- López-Andreo, M.; Garrido-Pertierra, A.; Puyet, A. Evaluation of post-polymerase chain reaction melting temperature analysis for meat species identification in mixed DNA samples. J. Agric. Food Chem. 2006, 54, 7973–7978. [Google Scholar] [CrossRef]
- Chuang, P.-S.; Hung, T.-C.; Chang, H.-A.; Huang, C.-K.; Shiao, J.-C. The species and origin of shark fins in Taiwan’s fishing ports, markets, and customs detention: A DNA barcoding analysis. PLoS ONE 2016, 11, e0147290. [Google Scholar] [CrossRef]
- Kastanos, E.; Papaneophytou, C.; Georgiou, T.; Demoliou, C. A simple and fast triplex-PCR for the identification of milk’s animal origin in Halloumi cheese and yoghurt. J. Dairy Res. 2022, 89, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Vankova, L.; Vanek, D. DNA-based identification of big cats and traditional Chinese medicine artifacts in the Czech Republic. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 122–124. [Google Scholar] [CrossRef]
- Votrubova, J.; Rihova, P.; Saskova, L.; Vanek, D. Operation Tiger’s Eye: DNA testing of traditional Chinese medicine artifacts in the Czech Republic. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e143–e144. [Google Scholar] [CrossRef]
- Gaubert, P.; Njiokou, F.; Olayemi, A.; Pagani, P.; Dufour, S.; Danquah, E.; Nutsuakor, M.E.K.; Ngua, G.; Missoup, A.D.; Tedesco, P.A. Bushmeat genetics: Setting up a reference framework for the DNA typing of A frican forest bushmeat. Mol. Ecol. Resour. 2015, 15, 633–651. [Google Scholar] [CrossRef]
- Lindsey, P.; Balme, G.; Becker, M.; Begg, C.; Bento, C.; Bocchino, C.; Zisadza, P. Illegal Hunting and the Bush-Meat Trade in Savanna Africa; Panthera, Zoological Society of London Wildlife Conservation Society: 2015. Available online: https://www.academia.edu/download/43979790/Illegal_hunting_and_the_bush-meat_trade_20160322-23875-iznbdi.pdf (accessed on 10 December 2025).
- Tikalova, E.; Votrubova, J.; Kufnerova, J.; Formanova, D.; Rihova, P.; Vankova, L.; Vanek, D. Busting the myths: DNA typeability after 48 hours of boil. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 79–82. [Google Scholar] [CrossRef]
- Hebenstreitova, K.; Salaba, O.; Trubac, J.; Kufnerova, J.; Vanek, D. The Influence of Tanning Chemical Agents on DNA Degradation: A Robust Procedure for the Analysis of Tanned Animal Hide—A Pilot Study. Life 2024, 14, 147. [Google Scholar] [CrossRef] [PubMed]
- Coghlan, M.L.; Haile, J.; Houston, J.; Murray, D.C.; White, N.E.; Moolhuijzen, P.; Bellgard, M.I.; Bunce, M. Deep sequencing of plant and animal DNA contained within traditional Chinese medicines reveals legality issues and health safety concerns. PLoS Genet. 2012, 8, e1002657. [Google Scholar] [CrossRef]
- Lance, R.F.; Guan, X. Variation in inhibitor effects on qPCR assays and implications for eDNA surveys. Can. J. Fish. Aquat. Sci. 2020, 77, 23–33. [Google Scholar] [CrossRef]
- Thompson, W.C.; Taroni, F.; Aitken, C.G. How the probability of a false positive affects the value of DNA evidence. J. Forensic Sci. 2003, 48, JFS2001171. [Google Scholar] [CrossRef]
- Galimberti, A.; Sandionigi, A.; Bruno, A.; Bellati, A.; Casiraghi, M. DNA barcoding in mammals: What’s new and where next? Hystrix 2015, 26, 13–24. [Google Scholar]
- Pálsdóttir, A.H.; Bläuer, A.; Rannamäe, E.; Boessenkool, S.; Hallsson, J.H. Not a limitless resource: Ethics and guidelines for destructive sampling of archaeofaunal remains. R. Soc. Open Sci. 2019, 6, 191059. [Google Scholar] [CrossRef]
- Brown, K.E. Developing Minimally Impactful Protocols for DNA Analysis of Museum Collection Bone Artifacts. Master’s Thesis, Simon Fraser University, Burnaby, BC, Canada, 2016. [Google Scholar]
- Polling, M.; Buij, R.; Laros, I.; de Groot, G.A. Continuous daily sampling of airborne eDNA detects all vertebrate species identified by camera traps. Environ. DNA 2024, 6, e591. [Google Scholar] [CrossRef]
- Zinger, L.; Benoiston, A.-S.; Cuenot, Y.; Leroy, C.; Louisanna, E.; Moreau, L.; Petitclerc, F.; Piatscheck, F.; Orivel, J.; Richard-Hansen, C. Elusive tropical forest canopy diversity revealed through environmental DNA contained in rainwater. Sci. Adv. 2025, 11, eadx4909. [Google Scholar] [CrossRef]
- Young, J.M.; Higgins, D.; Austin, J.J. Soil DNA: Advances in DNA technology offer a powerful new tool for forensic science. Geol. Soc. Lond. Spec. Publ. 2021, 492, 239–247. [Google Scholar] [CrossRef]
- Goray, M.; Taylor, D.; Bibbo, E.; Fantinato, C.; Fonneløp, A.E.; Gill, P.; van Oorschot, R.A. Emerging use of air eDNA and its application to forensic investigations—A review. Electrophoresis 2024, 45, 916–932. [Google Scholar] [CrossRef]
- Lewis, M.; Lainé, K.; Dawnay, L.; Lamont, D.; Scott, K.; Mariani, S.; Hänfling, B.; Dawnay, N. The forensic potential of environmental DNA (eDNA) in freshwater wildlife crime investigations: From research to application. Sci. Justice 2024, 64, 443–454. [Google Scholar] [CrossRef]
- Frøslev, T.G.; Ejrnæs, R.; Hansen, A.J.; Bruun, H.H.; Nielsen, I.B.; Ekelund, F.; Vestergård, M.; Kjøller, R. Treated like dirt: Robust forensic and ecological inferences from soil eDNA after challenging sample storage. Environ. DNA 2023, 5, 158–174. [Google Scholar] [CrossRef]
- Kelly, R.P.; Lodge, D.M.; Lee, K.N.; Theroux, S.; Sepulveda, A.J.; Scholin, C.A.; Craine, J.M.; Andruszkiewicz Allan, E.; Nichols, K.M.; Parsons, K.M. Toward a national eDNA strategy for the United States. Environ. DNA 2024, 6, e432. [Google Scholar] [CrossRef]
- LeClair, G.D.; Chatfield, M.W.; Kinnison, M.T. Environmental DNA as a tool for detecting illegal wildlife trade. Forensic Sci. Int. 2025, 370, 112446. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, C.B.; Keuschnig, C.; Larose, C.; Rissi, D.V.; Mourot, R.; Bradley, J.A.; Winkel, M.; Benning, L.G. DNA/RNA preservation in glacial snow and ice samples. Front. Microbiol. 2022, 13, 894893. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jeon, J.-Y.; Im, Y.-J.; Ha, N.; Kim, J.-K.; Moon, S.J.; Kim, M.-G. Long-term taxonomic and functional stability of the gut microbiome from human fecal samples. Sci. Rep. 2023, 13, 114. [Google Scholar] [CrossRef]
- Thraenhart, P.D.O.; Jursch, C. DNA/RNA Shield™. Available online: https://www.bioscience.co.uk/userfiles/pdf/Eurovir-test-report-DNA-RNA-Shield.pdf (accessed on 10 December 2025).
- Zhai, X.; Castro-Mejía, J.L.; Gobbi, A.; Aslampaloglou, A.; Kot, W.; Nielsen, D.S.; Deng, L. The impact of storage buffer and storage conditions on fecal samples for bacteriophage infectivity and metavirome analyses. Microbiome 2023, 11, 193. [Google Scholar] [CrossRef]
- Siembieda, J.L.; Miller, W.A.; Byrne, B.A.; Ziccardi, M.H.; Anderson, N.; Chouicha, N.; Sandrock, C.E.; Johnson, C.K. Zoonotic pathogens isolated from wild animals and environmental samples at two California wildlife hospitals. J. Am. Vet. Med. Assoc. 2011, 238, 773–783. [Google Scholar] [CrossRef]
- Smith, K.M.; Anthony, S.J.; Switzer, W.M.; Epstein, J.H.; Seimon, T.; Jia, H.; Sanchez, M.D.; Huynh, T.T.; Galland, G.G.; Shapiro, S.E. Zoonotic viruses associated with illegally imported wildlife products. PLoS ONE 2012, 7, e29505. [Google Scholar] [CrossRef]
- Dash, H.R.; Das, S. Microbial degradation of forensic samples of biological origin: Potential threat to human DNA typing. Mol. Biotechnol. 2018, 60, 141–153. [Google Scholar] [CrossRef] [PubMed]
- McCord, B.; Opel, K.; Funes, M.; Zoppis, S.; Meadows Jantz, L. An investigation of the effect of DNA degradation and inhibition on PCR amplification of single source and mixed forensic samples. In National Archive of Criminal Justice Data; US Department of Justice: Washington, DC, USA, 2011; pp. 1–65. Available online: https://iris.uniroma1.it/handle/11573/767000 (accessed on 1 September 2025).
- ISO 18385:2016; Minimizing the Risk of Human DNA Contamination in Products Used to Collect, Store and Analyze Biological Material for Forensic Purposes—Requirements. International Organization for Standardization (ISO): Geneva, Switzerland, 2016.
- Vanek, D.; Saskova, L.; Votrubova, J. Does the new ISO 18385: 2016 standard for forensic DNA-grade products need a revision? Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e148–e149. [Google Scholar] [CrossRef]
- Gutiérrez-Hurtado, I.A.; García-Acéves, M.E.; Puga-Carrillo, Y.; Guardado-Estrada, M.; Becerra-Loaiza, D.S.; Carrillo-Rodríguez, V.D.; Plazola-Zamora, R.; Godínez-Rubí, J.M.; Rangel-Villalobos, H.; Aguilar-Velázquez, J.A. Past, Present and Future Perspectives of Forensic Genetics. Biomolecules 2025, 15, 713. [Google Scholar] [CrossRef]
- Leonard, J.A.; Shanks, O.; Hofreiter, M.; Kreuz, E.; Hodges, L.; Ream, W.; Wayne, R.K.; Fleischer, R.C. Animal DNA in PCR reagents plagues ancient DNA research. J. Archaeol. Sci. 2007, 34, 1361–1366. [Google Scholar] [CrossRef]
- Czurda, S.; Smelik, S.; Preuner-Stix, S.; Nogueira, F.; Lion, T. Occurrence of fungal DNA contamination in PCR reagents: Approaches to control and decontamination. J. Clin. Microbiol. 2016, 54, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef]
- Champlot, S.; Berthelot, C.; Pruvost, M.; Bennett, E.A.; Grange, T.; Geigl, E.-M. An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS ONE 2010, 5, e13042. [Google Scholar] [CrossRef]
- Boessenkool, S.; Epp, L.S.; Haile, J.; Bellemain, E.; Edwards, M.; Coissac, E.; Willerslev, E.; Brochmann, C. Blocking human contaminant DNA during PCR allows amplification of rare mammal species from sedimentary ancient DNA. Mol. Ecol. 2012, 21, 1806–1815. [Google Scholar] [CrossRef]
- Huggins, L.G.; Koehler, A.V.; Schunack, B.; Inpankaew, T.; Traub, R.J. A host-specific blocking primer combined with optimal DNA extraction improves the detection capability of a metabarcoding protocol for canine vector-borne bacteria. Pathogens 2020, 9, 258. [Google Scholar] [CrossRef]
- Staats, M.; Arulandhu, A.J.; Gravendeel, B.; Holst-Jensen, A.; Scholtens, I.; Peelen, T.; Prins, T.W.; Kok, E. Advances in DNA metabarcoding for food and wildlife forensic species identification. Anal. Bioanal. Chem. 2016, 408, 4615–4630. [Google Scholar] [CrossRef]
- Mori, C.; Matsumura, S. Current issues for mammalian species identification in forensic science: A review. Int. J. Leg. Med. 2021, 135, 3–12. [Google Scholar] [CrossRef]
- Webster, L.M.; Prigge, T.-L.; Frankham, G.J. A guide for the validation of DNA based species identification in forensic casework. Forensic Sci. Int. Anim. Environ. 2024, 5, 100080. [Google Scholar] [CrossRef]
- Frankham, G.J.; Ogden, R.; Baker, B.W.; Ewart, K.M.; Johnson, R.N.; Kuiper, I.; Lindquist, C.D.; Moore, M.K.; Ndiaye, A.; Webster, L.M. Standards in wildlife forensic science, with a focus on non-human DNA analysis. Anim. Genet. 2025, 56, e70005. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.K.; Baker, B.W.; Bauman, T.L.; Burnham-Curtis, M.K.; Espinoza, E.O.; Ferrell, C.S.; Frankham, G.J.; Frazier, K.; Giles, J.L.; Hawk, D. The society for wildlife forensic science standards and guidelines. Forensic Sci. Int. Anim. Environ. 2021, 1, 100015. [Google Scholar] [CrossRef]
- Hochmeister, M.N.; Dirnhofer, R.; Borer, U.; Budowle, B.; Jung, J.; Comey, C.T. PCR-based typing of DNA extracted from cigarette butts. Int. J. Leg. Med. 1991, 104, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Chiou, F.S.; Pai, C.Y.; Hsu, Y.P.P.; Tsai, C.W.; Yang, C.H. Extraction of human DNA for PCR from chewed residues of betel quid using a novel “PVP/CTAB” method. J. Forensic Sci. 2001, 46, 1174–1179. [Google Scholar] [CrossRef]
- Volkmann, H.; Schwartz, T.; Kirchen, S.; Stofer, C.; Obst, U. Evaluation of inhibition and cross-reaction effects on real-time PCR applied to the total DNA of wastewater samples for the quantification of bacterial antibiotic resistance genes and taxon-specific targets. Mol. Cell. Probes 2007, 21, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Alderisio, K.A.; Singh, A.; Xiao, L. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 2005, 71, 1135–1141. [Google Scholar] [CrossRef]
- Braid, M.D.; Daniels, L.M.; Kitts, C.L. Removal of PCR inhibitors from soil DNA by chemical flocculation. J. Microbiol. Methods 2003, 52, 389–393. [Google Scholar] [CrossRef]
- Wang, H.; Qi, J.; Xiao, D.; Wang, Z.; Tian, K. A re-evaluation of dilution for eliminating PCR inhibition in soil DNA samples. Soil Biol. Biochem. 2017, 106, 109–118. [Google Scholar] [CrossRef]
- Vankova, L.; Alaverdyan, J.; Vanek, D. Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping. Life 2025, 15, 664. [Google Scholar] [CrossRef] [PubMed]
- Pruvost, M.; Geigl, E.-M. Real-time quantitative PCR to assess the authenticity of ancient DNA amplification. J. Archaeol. Sci. 2004, 31, 1191–1197. [Google Scholar] [CrossRef]
- Mahlerová, K.; Alaverdyan, J.; Vaňková, L.; Vaněk, D. Molecular Tools for Lynx spp. qPCR Identification and STR-Based Individual Identification of Eurasian Lynx (Lynx lynx) in Forensic Casework. Methods Protoc. 2025, 8, 47. [Google Scholar] [CrossRef]
- Hebenstreitová, K.; Vaňková, L.; Vaněk, D. Determination of the suitability of agilent bioanalyzer 2100 for investigations into wildlife crimes: Case studies. Eur. J. Environ. Sci. 2025, 15, 43–52. [Google Scholar] [CrossRef]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 42. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. London. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Sharma, P.; Kobayashi, T. Are “universal” DNA primers really universal? J. Appl. Genet. 2014, 55, 485–496. [Google Scholar] [CrossRef]
- Mioduchowska, M.; Czyż, M.J.; Gołdyn, B.; Kur, J.; Sell, J. Instances of erroneous DNA barcoding of metazoan invertebrates: Are universal cox1 gene primers too “universal”? PLoS ONE 2018, 13, e0199609. [Google Scholar] [CrossRef]
- Phillips, J.D.; Gillis, D.J.; Hanner, R.H. Incomplete estimates of genetic diversity within species: Implications for DNA barcoding. Ecol. Evol. 2019, 9, 2996–3010. [Google Scholar] [CrossRef] [PubMed]
- Ewart, K.M.; Lightson, A.L.; Sitam, F.T.; Rovie-Ryan, J.; Nguyen, S.G.; Morgan, K.I.; Luczon, A.; Anadon, E.M.S.; De Bruyn, M.; Bourgeois, S. DNA analyses of large pangolin scale seizures: Species identification validation and case studies. Forensic Sci. Int. Anim. Environ. 2021, 1, 100014. [Google Scholar] [CrossRef]
- Brown, A.O.; Ueland, M.; Stuart, B.H.; Frankham, G.J. A forensically validated genetic toolkit for the species and lineage identification of the highly trafficked shingleback lizard (Tiliqua rugosa). Forensic Sci. Int. Genet. 2023, 62, 102784. [Google Scholar] [CrossRef]
- Hatten, C.E.; Fitriana, Y.S.; Prigge, T.-L.; Irham, M.; Sutrisno, H.; Dingle, C. DNA analysis and validation for species identification of seized helmeted hornbill (Rhinoplax vigil) casques. Forensic Sci. Int. Anim. Environ. 2023, 3, 100058. [Google Scholar] [CrossRef]
- Matthes, N.; Pietsch, K.; Rullmann, A.; Näumann, G.; Pöpping, B.; Szabo, K. The Barcoding Table of Animal Species (BaTAnS): A new tool to select appropriate methods for animal species identification using DNA barcoding. Mol. Biol. Rep. 2020, 47, 6457–6461. [Google Scholar] [CrossRef] [PubMed]
- Schröder, U.; Sotelo, C.G.; Klapper, R. FISH-FIT: A web-based tool to improve European seafood authenticity control. Food Control 2025, 176, 111335. [Google Scholar] [CrossRef]
- Hsieh, H.-M.; Chiang, H.-L.; Tsai, L.-C.; Lai, S.-Y.; Huang, N.-E.; Linacre, A.; Lee, J.C.-I. Cytochrome b gene for species identification of the conservation animals. Forensic Sci. Int. 2001, 122, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Crossley, B.M.; Bai, J.; Glaser, A.; Maes, R.; Porter, E.; Killian, M.L.; Clement, T.; Toohey-Kurth, K. Guidelines for Sanger sequencing and molecular assay monitoring. J. Vet. Diagn. Investig. 2020, 32, 767–775. [Google Scholar] [CrossRef]
- Linacre, A.; Lee, J.C.-I. Species determination: The role and use of the cytochrome b gene. In Forensic DNA Typing Protocols; Springer: Berlin/Heidelberg, Germany, 2016; pp. 287–296. [Google Scholar]
- Wilkinson, M.J.; Szabo, C.; Ford, C.S.; Yarom, Y.; Croxford, A.E.; Camp, A.; Gooding, P. Replacing Sanger with Next Generation Sequencing to improve coverage and quality of reference DNA barcodes for plants. Sci. Rep. 2017, 7, srep46040. [Google Scholar] [CrossRef]
- Illumina. NovaSeq6000 Sequencing System Guide. Available online: https://support.illumina.com/content/dam/illumina-support/documents/documentation/system_documentation/novaseq/1000000019358_18_novaseq-6000-system-guide.pdf (accessed on 7 October 2025).
- Domrazek, K.; Jurka, P. Application of next-generation sequencing (NGS) techniques for selected companion animals. Animals 2024, 14, 1578. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Ghionda, M.C.; D’Alessandro, E.; Geraci, C.; Chiofalo, V.; Fontanesi, L. A next generation semiconductor based sequencing approach for the identification of meat species in DNA mixtures. PLoS ONE 2015, 10, e0121701. [Google Scholar] [CrossRef]
- Seah, A.; Lim, M.C.; McAloose, D.; Prost, S.; Seimon, T.A. MinION-based DNA barcoding of preserved and non-invasively collected wildlife samples. Genes 2020, 11, 445. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Fuselli, S.; Baptista, R.; Panziera, A.; Magi, A.; Guglielmi, S.; Tonin, R.; Benazzo, A.; Bauzer, L.; Mazzoni, C.; Bertorelle, G. A new hybrid approach for MHC genotyping: High-throughput NGS and long read MinION nanopore sequencing, with application to the non-model vertebrate Alpine chamois (Rupicapra rupicapra). Heredity 2018, 121, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Menegon, M.; Cantaloni, C.; Rodriguez-Prieto, A.; Centomo, C.; Abdelfattah, A.; Rossato, M.; Bernardi, M.; Xumerle, L.; Loader, S.; Delledonne, M. On site DNA barcoding by nanopore sequencing. PLoS ONE 2017, 12, e0184741. [Google Scholar] [CrossRef] [PubMed]
- Sim, J.; Chapman, B. In-field whole genome sequencing using the MinION nanopore sequencer to detect the presence of high-prized military targets. Aust. J. Forensic Sci. 2019, 51, S86–S90. [Google Scholar] [CrossRef]
- Tyler, A.D.; McAllister, J.; Stapleton, H.; Gauci, P.; Antonation, K.; Thirkettle-Watts, D.; Corbett, C.R. Field-based detection of bacteria using nanopore sequencing: Method evaluation for biothreat detection in complex samples. PLoS ONE 2023, 18, e0295028. [Google Scholar] [CrossRef]
- Mahlerová, K.; Vaňková, L.; Tomsia, M.; Vaněk, D. Rapid Species Barcoding Using Bento Lab Mobile Laboratory. Forensic Sci. 2024, 4, 566–572. [Google Scholar] [CrossRef]
- Mueller, S.; Handy, S.M.; Deeds, J.R.; George, G.O.; Broadhead, W.J.; Pugh, S.E.; Garrett, S.D. Development of a COX1 based PCR-RFLP method for fish species identification. Food Control 2015, 55, 39–42. [Google Scholar] [CrossRef]
- Denyingyhot, A.; Phraephaisarn, C.; Vesaratchavest, M.; Dahlan, W.; Keeratipibul, S. A new tool for quality control to monitor contamination of six non-halal meats in food industry by multiplex high-resolution melting analysis (HRMA). NFS J. 2021, 25, 31–40. [Google Scholar] [CrossRef]
- Friedenberger, A.; Doyle, C.; Couillard, L.; Kyle, C.J. The bear necessities: A sensitive qPCR assay for bear DNA detection from bile and derived products to complement wildlife forensic enforcement. Forensic Sci. Int. Genet. 2023, 67, 102935. [Google Scholar] [CrossRef]
- Lahiff, S.; Glennon, M.; Lyng, J.; Smith, T.; Maher, M.; Shilton, N. Species-specific PCR for the identification of ovine, porcine and chicken species in meat and bone meal (MBM). Mol. Cell. Probes 2001, 15, 27–35. [Google Scholar] [CrossRef]
- Noikotr, K.; Chaveerach, A.; Pinthong, K.; Tanomtong, A.; Sudmoon, R.; Tanee, T. RAPD and barcode analyses of groupers of the genus Epinephelus. Genet. Mol. Res 2013, 12, 5721–5732. [Google Scholar] [CrossRef]
- Wood, S.A.; Pochon, X.; Laroche, O.; von Ammon, U.; Adamson, J.; Zaiko, A. A comparison of droplet digital polymerase chain reaction (PCR), quantitative PCR and metabarcoding for species-specific detection in environmental DNA. Mol. Ecol. Resour. 2019, 19, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Basanisi, M.G.; La Bella, G.; Nobili, G.; Coppola, R.; Damato, A.M.; Cafiero, M.A.; La Salandra, G. Application of the novel Droplet digital PCR technology for identification of meat species. Int. J. Food Sci. Technol. 2020, 55, 1145–1150. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, S.E.; Ko, K.S.; Park, S.H. Identification of forensically important Calliphoridae and Sarcophagidae species collected in Korea using SNaPshot multiplex system targeting the cytochrome c oxidase subunit i gene. BioMed Res. Int. 2018, 2018, 2953892. [Google Scholar] [CrossRef]
- Hoffman, J.; Clark, M.; Amos, W.; Peck, L. Widespread amplification of amplified fragment length polymorphisms (AFLPs) in marine Antarctic animals. Polar Biol. 2012, 35, 919–929. [Google Scholar] [CrossRef]
- Mori, C.; Matsumura, S. Development and validation of simultaneous identification of 26 mammalian and poultry species by a multiplex assay. Int. J. Leg. Med. 2021, 136, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Carneiro, J.; Matthiesen, R.; van Asch, B.; Pinto, N.; Gusmao, L.; Amorim, A. Identification of species by multiplex analysis of variable-length sequences. Nucleic Acids Res. 2010, 38, e203. [Google Scholar] [CrossRef]
- Pun, K.M.; Albrecht, C.; Castella, V.; Fumagalli, L. Species identification in mammals from mixed biological samples based on mitochondrial DNA control region length polymorphism. Electrophoresis 2009, 30, 1008–1014. [Google Scholar] [CrossRef]
- Vankova, L.; Vanek, D. Capillary-Electrophoresis-Based species barcoding of big cats: CR-mtDNA-Length Polymorphism. Life 2024, 14, 497. [Google Scholar] [CrossRef]
- El Sheikha, A.F.; Mokhtar, N.F.K.; Amie, C.; Lamasudin, D.U.; Isa, N.M.; Mustafa, S. Authentication technologies using DNA-based approaches for meats and halal meats determination. Food Biotechnol. 2017, 31, 281–315. [Google Scholar] [CrossRef]
- Hossain, M.M.; Uddin, S.M.K.; Sultana, S.; Wahab, Y.A.; Sagadevan, S.; Johan, M.R.; Ali, M.E. Authentication of Halal and Kosher meat and meat products: Analytical approaches, current progresses and future prospects. Crit. Rev. Food Sci. Nutr. 2021, 62, 285–310. [Google Scholar] [CrossRef]
- Wang, N.; Xing, R.-R.; Zhou, M.-Y.; Sun, R.-X.; Han, J.-X.; Zhang, J.-K.; Zheng, W.-J.; Chen, Y. Application of DNA barcoding and metabarcoding for species identification in salmon products. Food Addit. Contam. Part A 2021, 38, 754–768. [Google Scholar] [CrossRef]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-X.; Hewitt, G.M. Nuclear integrations: Challenges for mitochondrial DNA markers. Trends Ecol. Evol. 1996, 11, 247–251. [Google Scholar] [CrossRef]
- Muskavitch, K.M.T.; Linn, S. 13 recBC-like Enzymes: Exonuclease V Deoxyribonucleases. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 1981; Volume 14, pp. 233–250. [Google Scholar]
- Morgan, K.I.; Ewart, K.M.; Nguyen, T.Q.; Sitam, F.T.; Ouitavon, K.; Lightson, A.L.; Kotze, A.; McEwing, R. Avoiding common numts to provide reliable species identification for tiger parts. Forensic Sci. Int. Rep. 2021, 3, 100166. [Google Scholar] [CrossRef]
- Wolff, J.N.; Shearman, D.C.; Brooks, R.C.; Ballard, J.W. Selective enrichment and sequencing of whole mitochondrial genomes in the presence of nuclear encoded mitochondrial pseudogenes (numts). PLoS ONE 2012, 7, e37142. [Google Scholar] [CrossRef]
- Lascaro, D.; Castellana, S.; Gasparre, G.; Romeo, G.; Saccone, C.; Attimonelli, M. The RHNumtS compilation: Features and bioinformatics approaches to locate and quantify Human NumtS. BMC Genom. 2008, 9, 267. [Google Scholar] [CrossRef]
- Ring, J.D.; Sturk-Andreaggi, K.; Alyse Peck, M.; Marshall, C. Bioinformatic removal of NUMT-associated variants in mitotiling next-generation sequencing data from whole blood samples. Electrophoresis 2018, 39, 2785–2797. [Google Scholar] [CrossRef]
- Cihlar, J.C.; Strobl, C.; Lagacé, R.; Muenzler, M.; Parson, W.; Budowle, B. Distinguishing mitochondrial DNA and NUMT sequences amplified with the precision ID mtDNA whole genome panel. Mitochondrion 2020, 55, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Gabryś, J.; Kij, B.; Kochan, J.; Bugno-Poniewierska, M. Interspecific hybrids of animals-in nature, breeding and science—A review. Ann. Anim. Sci. 2021, 21, 403–415. [Google Scholar] [CrossRef]
- Amorim, A.; Pereira, F.; Alves, C.; García, O. Species assignment in forensics and the challenge of hybrids. Forensic Sci. Int. Genet. 2020, 48, 102333. [Google Scholar] [CrossRef]
- Rastogi, G.; Dharne, M.S.; Walujkar, S.; Kumar, A.; Patole, M.S.; Shouche, Y.S. Species identification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 2007, 76, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Peltola, T.; Heikkilä, J. Outlaws or protected? DNA, hybrids, and biopolitics in a Finnish wolf-poaching case. Soc. Anim. 2018, 26, 197–216. [Google Scholar] [CrossRef]
- Vonholdt, B.M.; Pollinger, J.P.; Earl, D.A.; Parker, H.G.; Ostrander, E.A.; Wayne, R.K. Identification of recent hybridization between gray wolves and domesticated dogs by SNP genotyping. Mamm. Genome 2013, 24, 80–88. [Google Scholar] [CrossRef]
- Jiang, H.H.; Li, B.; Ma, Y.; Bai, S.Y.; Dahmer, T.D.; Linacre, A.; Xu, Y.C. Forensic validation of a panel of 12 SNPs for identification of Mongolian wolf and dog. Sci. Rep. 2020, 10, 13249. [Google Scholar] [CrossRef]
- Lorenzini, R.; Attili, L.; Tancredi, C.; Fanelli, R.; Garofalo, L. A validated molecular protocol to differentiate pure wolves, dogs and wolf x dog hybrids through a panel of multiplexed canine STR markers. Diversity 2022, 14, 511. [Google Scholar] [CrossRef]
- Kwong, S.; Srivathsan, A.; Meier, R. An update on DNA barcoding: Low species coverage and numerous unidentified sequences. Cladistics 2012, 28, 639–644. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef]
- Consortium, D.T.o.L.P. Sequence locally, think globally: The Darwin Tree of Life Project. Proc. Natl. Acad. Sci. USA 2022, 119, e2115642118. [Google Scholar]
- Twyford, A.D.; Beasley, J.; Barnes, I.; Allen, H.; Azzopardi, F.; Bell, D.; Blaxter, M.L.; Broad, G.; Campos-Dominguez, L.; Choonea, D. A DNA barcoding framework for taxonomic verification in the Darwin Tree of Life Project. Wellcome Open Res. 2024, 9, 339. [Google Scholar] [CrossRef]
- Jones, L.; Twyford, A.D.; Ford, C.R.; Rich, T.C.; Davies, H.; Forrest, L.L.; Hart, M.L.; McHaffie, H.; Brown, M.R.; Hollingsworth, P.M. Barcode UK: A complete DNA barcoding resource for the flowering plants and conifers of the United Kingdom. Mol. Ecol. Resour. 2021, 21, 2050–2062. [Google Scholar] [CrossRef]
- Janzen, D.; Hallwachs, W. How a tropical country can DNA barcode itself. IBOL Barcode Bull. 2019, 9. Available online: https://journal.lib.uoguelph.ca/index.php/ibol/article/download/5526/5454 (accessed on 10 December 2025).
- Pentinsaari, M. Utility of DNA Barcodes in Identification and Delimitation of Beetle Species, with Insights into COI Protein Structure Across the Animal Kingdom; University of Oulu: Oulu, Finland, 2016. [Google Scholar]
- Zangl, L.; Daill, D.; Schweiger, S.; Gassner, G.; Koblmueller, S. A reference DNA barcode library for Austrian amphibians and reptiles. PLoS ONE 2020, 15, e0229353. [Google Scholar] [CrossRef]
- Zúñiga, J.D.; Gostel, M.R.; Mulcahy, D.G.; Barker, K.; Hill, A.; Sedaghatpour, M.; Vo, S.Q.; Funk, V.A.; Coddington, J.A. Data release: DNA barcodes of plant species collected for the Global Genome Initiative for Gardens program, National Museum of Natural History, Smithsonian Institution. PhytoKeys 2017, 88, 119. [Google Scholar] [CrossRef] [PubMed]
- Queirós, J.; Silva, R.; Pinho, C.J.; Vale-Gonçalves, H.M.; Pita, R.; Alves, P.C.; Beja, P.; Paupério, J.; Porto, M. The InBIO Barcoding Initiative Database: Contribution to the knowledge of DNA barcodes of the vascular plants of north-eastern Portugal. Biodivers. Data J. 2025, 13, e142020. [Google Scholar] [CrossRef] [PubMed]
- Chenery, J. Launch of International Barcode of Life Project activates world’s largest biodiversity genomics initiative. Biopreserv. Biobank. 2010, 8, S11. [Google Scholar]
- Ahlers, N.; Creecy, J.; Frankham, G.; Johnson, R.; Kotze, A.; Linacre, A.; McEwing, R.; Mwale, M.; Rovie-Ryan, J.; Sitam, F. ‘ForCyt’DNA database of wildlife species. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e466–e468. [Google Scholar] [CrossRef]
- Au, W.C.; Dures, S.G.; Ishida, Y.; Green, C.E.; Zhao, K.; Ogden, R.; Roca, A.L. Lion Localizer: A software tool for inferring the provenance of lions (Panthera leo) using mitochondrial DNA. J. Hered. 2024, 115, 166–172. [Google Scholar] [CrossRef]
- Zhao, K.; Ishida, Y.; Green, C.E.; Davidson, A.G.; Sitam, F.A.; Donnelly, C.L.; De Flamingh, A.; Perrin-Stowe, T.I.; Bourgeois, S.; Brandt, A.L. Loxodonta Localizer: A software tool for inferring the provenance of African elephants and their ivory using mitochondrial DNA. J. Hered. 2019, 110, 761–768. [Google Scholar] [CrossRef]
- Nakazato, T. Survey of species covered by DNA barcoding data in BOLD and GenBank for integration of data for museomics. Biodivers. Inf. Sci. Stand. 2020, 4, e59065. [Google Scholar] [CrossRef]
- Baena-Bejarano, N.; Reina, C.; Martínez-Revelo, D.E.; Medina, C.A.; Tovar, E.; Uribe-Soto, S.; Neita-Moreno, J.C.; Gonzalez, M.A. Taxonomic identification accuracy from BOLD and GenBank databases using over a thousand insect DNA barcodes from Colombia. PLoS ONE 2023, 18, e0277379. [Google Scholar] [CrossRef] [PubMed]
- Meiklejohn, K.A.; Damaso, N.; Robertson, J.M. Assessment of BOLD and GenBank–Their accuracy and reliability for the identification of biological materials. PLoS ONE 2019, 14, e0217084. [Google Scholar] [CrossRef] [PubMed]
- Bridge, P.D.; Roberts, P.J.; Spooner, B.M.; Panchal, G. On the unreliability of published DNA sequences. New Phytol. 2003, 160, 43–48. [Google Scholar] [CrossRef]
- Jin, S.; Kim, K.Y.; Kim, M.-S.; Park, C. An assessment of the taxonomic reliability of DNA barcode sequences in publicly available databases. Algae 2020, 35, 293–301. [Google Scholar] [CrossRef]
- Kjærandsen, J. Current state of DNA barcoding of Sciaroidea (Diptera)—Highlighting the need to build the reference library. Insects 2022, 13, 147. [Google Scholar] [CrossRef]
- Patel, K.K.; Scheible, M.K.; Meiklejohn, K.A. Interlaboratory study to assess the practical utility of OSAC proposed standard 2021-S-0006: Standard for the use of GenBank for taxonomic assignment of wildlife. Forensic Sci. Int. Anim. Environ. 2023, 4, 100067. [Google Scholar] [CrossRef]
- DNA Ties Canada to U.S. Mad Cow. Available online: https://www.cbsnews.com/news/dna-ties-canada-to-us-mad-cow/ (accessed on 2 September 2025).
- DNA Tests Show U.S. Cow with Mad Cow Disease Came from Canada. Available online: https://www.pbs.org/newshour/health/health-jan-june04-madcow_01-06 (accessed on 2 September 2025).
- Harper, C.K. RhODIS® (The Rhinoceros DNA Index System): The Application of Simple Forensic and Genetic Tools Help Conserve African Rhinoceros. In Wildlife Biodiversity Conservation; Springer: Berlin/Heidelberg, Germany, 2021; pp. 463–485. [Google Scholar]
- Harper, C.; Ludwig, A.; Clarke, A.; Makgopela, K.; Yurchenko, A.; Guthrie, A.; Dobrynin, P.; Tamazian, G.; Emslie, R.; Van Heerden, M. Robust forensic matching of confiscated horns to individual poached African rhinoceros. Curr. Biol. 2018, 28, R13–R14. [Google Scholar] [CrossRef]
- Singh, A.; Priyambada, P.; Jabin, G.; Singh, S.K.; Joshi, B.D.; Venkatraman, C.; Chandra, K.; Sharma, L.K.; Thakur, M. Pangolin Indexing System: Implications in forensic surveillance of large seizures. Int. J. Leg. Med. 2020, 134, 1613–1618. [Google Scholar] [CrossRef] [PubMed]
- Roberto, B.; Mauro, Z.; Claudia, C.; Gianluca, D.; Marta, B.; Michel, D.; Luciano, D.T.; Luigi, L.F.; Oliviero, O.; Francesco, P. Who’s who in the western Hermann’s tortoise conservation: A STR toolkit and reference database for wildlife forensic genetic analyses. bioRxiv 2018, 484030. Available online: https://www.biorxiv.org/content/10.1101/484030.abstract (accessed on 10 December 2025).
- Biello, R.; Zampiglia, M.; Corti, C.; Deli, G.; Biaggini, M.; Crestanello, B.; Delaugerre, M.; Di Tizio, L.; Leonetti, F.L.; Casari, S. Mapping the geographic origin of captive and confiscated Hermann’s tortoises: A genetic toolkit for conservation and forensic analyses. Forensic Sci. Int. Genet. 2021, 51, 102447. [Google Scholar] [CrossRef]
- Jan, C.; Fumagalli, L. Polymorphic DNA microsatellite markers for forensic individual identification and parentage analyses of seven threatened species of parrots (family Psittacidae). PeerJ 2016, 4, e2416. [Google Scholar] [CrossRef]
- Willows-Munro, S.; Kleinhans, C. Testing microsatellite loci for individual identification of captive African grey parrots (Psittacus erithacus): A molecular tool for parentage analysis that will aid in monitoring legal trade. Conserv. Genet. Resour. 2020, 12, 489–495. [Google Scholar] [CrossRef]
- De Bruyn, M.; Dalton, D.L.; Mwale, M.; Ehlers, K.; Kotze, A. Development and Validation of a Novel Forensic STR Multiplex Assay for Blue (Anthropoides paradiseus), Wattled (Bugeranus carunculatus), and Grey-Crowned Crane (Balearica regulorum). Forensic Sci. Int. Genet. 2024, 73, 103100. [Google Scholar] [CrossRef]
- Vaněk, D.; Ehler, E.; Vaňková, L. Development of DNA quantitation and STR typing systems for Panthera tigris species determination and individual identification in forensic casework. Eur. J. Environ. Sci. 2021, 11, 113–118. [Google Scholar] [CrossRef]
- Ewart, K.M.; Sitam, F.T.; Ali, N.A.N.B.G.; Ogden, R.; Morgan, K.I.; Tran, H.M.; Bui, T.P.; Nguyen, T.Q.; Nguyen, S.G.; Rosli, N. TigerBase: A DNA registration system to enhance enforcement and compliance testing of captive tiger facilities. Forensic Sci. Int. Genet. 2025, 74, 103149. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.-Z.; Du, L.-M.; Yang, B.; Shen, F.-J.; Zhang, H.-M.; Zhang, Z.-H.; Zhang, X.-Y.; Yue, B.-S. Genome-wide survey and analysis of microsatellites in giant panda (Ailuropoda melanoleuca), with a focus on the applications of a novel microsatellite marker system. BMC Genom. 2015, 16, 61. [Google Scholar] [CrossRef]
- Xu, W.; Jiang, X.; Chen, M.; Xie, D.; Tang, L.; Wang, W.; Zhang, X.; Shen, F. Development of a panel of microhaplotype markers for giant panda. Anim. Genet. 2025, 56, e70008. [Google Scholar] [CrossRef]
- Potoczniak, M.J.; Chermak, M.; Quarino, L.; Tobe, S.S.; Conte, J. Development of a multiplex, PCR-based genotyping assay for African and Asian elephants for forensic purposes. Int. J. Leg. Med. 2020, 134, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Kinuthia, J.; Harper, C.; Muya, S.; Kimwele, C.; Alakonya, A.; Muigai, A.; Gakuya, F.; Mwaniki, M.; Gatebe, E. The selection of a standard STR panel for DNA profiling of the African elephant (Loxodonta africana) in Kenya. Conserv. Genet. Resour. 2015, 7, 305–307. [Google Scholar] [CrossRef]
- Gutiérrez-Rodríguez, J.; Zaldívar-Riverón, A.; Solano-Zavaleta, I.; Campbell, J.A.; Meza-Lázaro, R.N.; Flores-Villela, O.; de Oca, A.N.-M. Phylogenomics of the Mesoamerican alligator-lizard genera Abronia and Mesaspis (Anguidae: Gerrhonotinae) reveals multiple independent clades of arboreal and terrestrial species. Mol. Phylogenet. Evol. 2021, 154, 106963. [Google Scholar] [CrossRef] [PubMed]
- Hale, M.L.; Burg, T.M.; Steeves, T.E. Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 2012, 7, e45170. [Google Scholar] [CrossRef]
- Sajantila, A.; Lukka, M.; Syvänen, A.-C. Experimentally observed germline mutations at human micro-and minisatellite loci. Eur. J. Hum. Genet. 1999, 7, 263–266. [Google Scholar] [CrossRef]
- Renner, S.C.; Neumann, D.; Burkart, M.; Feit, U.; Giere, P.; Gröger, A.; Paulsch, A.; Paulsch, C.; Sterz, M.; Vohland, K. Import and export of biological samples from tropical countries–considerations and guidelines for research teams. Org. Divers. Evol. 2012, 12, 81–98. [Google Scholar] [CrossRef]
- Chege, M.; Sewalt, B.; Lesilau, F.; de Snoo, G.; Patterson, B.D.; Kariuki, L.; Otiende, M.; Omondi, P.; De Iongh, H.; Vrieling, K. Genetic diversity of lion populations in Kenya: Evaluating past management practices and recommendations for future conservation actions. Evol. Appl. 2024, 17, e13676. [Google Scholar] [CrossRef]
- Radko, A.; Podbielska, A. Microsatellite DNA analysis of genetic diversity and parentage testing in the popular dog breeds in Poland. Genes 2021, 12, 485. [Google Scholar] [CrossRef]
- Prajapati, S. Consanguine Marriage Leads to Hierarchical Imbalance of ABO and STR Frequency and Affects the Genetic Diversity. Curr. Forensic Sci. 2025, 3, E26664844350819. [Google Scholar] [CrossRef]
- Parra, G.J.; Cagnazzi, D.; Jedensjö, M.; Ackermann, C.; Frere, C.; Seddon, J.; Nikolic, N.; Krützen, M. Low genetic diversity, limited gene flow and widespread genetic bottleneck effects in a threatened dolphin species, the Australian humpback dolphin. Biol. Conserv. 2018, 220, 192–200. [Google Scholar] [CrossRef]
- Gustafson, K.D.; Hawkins, M.G.; Drazenovich, T.L.; Church, R.; Brown, S.A.; Ernest, H.B. Founder events, isolation, and inbreeding: Intercontinental genetic structure of the domestic ferret. Evol. Appl. 2018, 11, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Matocq, M.D.; Villablanca, F.X. Low genetic diversity in an endangered species: Recent or historic pattern? Biol. Conserv. 2001, 98, 61–68. [Google Scholar] [CrossRef]
- Bristol, R.M.; Tucker, R.; Dawson, D.A.; Horsburgh, G.; Prys-Jones, R.P.; Frantz, A.C.; Krupa, A.; Shah, N.J.; Burke, T.; Groombridge, J.J. Comparison of historical bottleneck effects and genetic consequences of re-introduction in a critically endangered island passerine. Mol. Ecol. 2013, 22, 4644–4662. [Google Scholar] [CrossRef]
- Ge, J.; Eisenberg, A.; Budowle, B. Developing criteria and data to determine best options for expanding the core CODIS loci. Investig. Genet. 2012, 3, 1. [Google Scholar] [CrossRef]
- Budowle, B.; Moretti, T.R.; Niezgoda, S.J.; Brown, B.L. CODIS and PCR-based short tandem repeat loci: Law enforcement tools. In Second European Symposium on Human Identification; Promega Corporation: Madison, WI, USA, 1998; pp. 73–88. [Google Scholar]
- Ge, J.; Budowle, B. Forensic investigation approaches of searching relatives in DNA databases. J. Forensic Sci. 2021, 66, 430–443. [Google Scholar] [CrossRef]
- Tigris ID Database. Available online: https://cites.org/sites/default/files/eng/prog/enforcement/TIGER%20GENETICS-Project%20TigrisID.pdf (accessed on 23 August 2025).
- Wasser, S.K.; Shedlock, A.M.; Comstock, K.; Ostrander, E.A.; Mutayoba, B.; Stephens, M. Assigning African elephant DNA to geographic region of origin: Applications to the ivory trade. Proc. Natl. Acad. Sci. USA 2004, 101, 14847–14852. [Google Scholar] [CrossRef] [PubMed]
- Wasser, S.K.; Wolock, C.J.; Kuhner, M.K.; Brown, J.E., III; Morris, C.; Horwitz, R.J.; Wong, A.; Fernandez, C.J.; Otiende, M.Y.; Hoareau, Y. Elephant genotypes reveal the size and connectivity of transnational ivory traffickers. Nat. Hum. Behav. 2022, 6, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Halverson, J.; Basten, C. A PCR multiplex and database for forensic DNA identification of dogs. J. Forensic Sci. 2005, 50, JFS2004207–JFS2004212. [Google Scholar] [CrossRef]
- Kanthaswamy, S.; Tom, B.K.; Mattila, A.M.; Johnston, E.; Dayton, M.; Kinaga, J.; Joy-Alise Erickson, B.; Halverson, J.; Fantin, D.; DeNise, S. Canine population data generated from a multiplex STR kit for use in forensic casework. J. Forensic Sci. 2009, 54, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Kun, T.J. Development and Validation of a Canine MiniSTR Panel for Forensic Casework; University of California: Davis, CA, USA, 2011. [Google Scholar]
- Menotti-Raymond, M.; David, V.A.; Weir, B.S.; O’Brien, S.J. A population genetic database of cat breeds developed in coordination with a domestic cat STR multiplex. J. Forensic Sci. 2012, 57, 596–601. [Google Scholar] [CrossRef]
- Ivanković, A.; Bittante, G.; Konjačić, M.; Kelava Ugarković, N.; Pećina, M.; Ramljak, J. Evaluation of the conservation status of the Croatian Posavina horse breed based on pedigree and microsatellite data. Animals 2021, 11, 2130. [Google Scholar] [CrossRef]
- Tobe, S.S.; Reid, S.J.; Linacre, A.M. Successful DNA typing of a drug positive urine sample from a race horse. Forensic Sci. Int. 2007, 173, 85–86. [Google Scholar] [CrossRef]
- Parson, W.; Dür, A. EMPOP—A forensic mtDNA database. Forensic Sci. Int. Genet. 2007, 1, 88–92. [Google Scholar] [CrossRef]
- Willuweit, S.; Roewer, L.; International Forensic Y Chromosome User Group. Y chromosome haplotype reference database (YHRD): Update. Forensic Sci. Int. Genet. 2007, 1, 83–87. [Google Scholar] [CrossRef]
- ISO/IEC 17025:2017; General Requirements for the Competence of Testing and Calibration Laboratories. International Organization for Standardization (ISO)/International Electrotechnical Commission (IEC): Geneva, Switzerland, 2017.
- ISO 21043; Forensic Sciences (This is a Multi-Part Standard with Different Publication Years for Each Part). International Organization for Standardization (ISO): Geneva, Switzerland, 2025.
- Morrison, G.S.; Elliott, S.; Guiness, J.; Sonden, L.; Court, D.S. A guide to ISO 21043 forensic sciences from the perspective of the forensic-data-science paradigm. Sci. Justice 2025, 65, 101304. [Google Scholar] [CrossRef]
- Berger, C.E. Finally a really forensic worldwide standard: ISO 21043 Forensic sciences, Part 4, Interpretation. Forensic Sci. Int. Synerg. 2025, 10, 100589. [Google Scholar] [CrossRef]
- Alves, C.; Pereira, R.; Prieto, L.; Aler, M.; Amaral, C.R.; Arévalo, C.; Berardi, G.; Di Rocco, F.; Caputo, M.; Carmona, C.H. Species identification in forensic samples using the SPInDel approach: A GHEP-ISFG inter-laboratory collaborative exercise. Forensic Sci. Int. Genet. 2017, 28, 219–224. [Google Scholar] [CrossRef]
- Ewart, K.M.; Frankham, G.J.; McEwing, R.; Webster, L.M.; Ciavaglia, S.A.; Linacre, A.M.; The, D.T.; Ovouthan, K.; Johnson, R.N. An internationally standardized species identification test for use on suspected seized rhinoceros horn in the illegal wildlife trade. Forensic Sci. Int. Genet. 2018, 32, 33–39. [Google Scholar] [CrossRef] [PubMed]
- McEwing, R.; Ewart, K.M.; Morgan, K.I. A simple procedure for mimicking tissue samples from CITES controlled animal species for use in DNA proficiency tests. Forensic Sci. Int. Rep. 2021, 3, 100213. [Google Scholar] [CrossRef]
- Ueland, M.; Brown, A.; Bartos, C.; Frankham, G.J.; Johnson, R.N.; Forbes, S.L. Profiling volatilomes: A novel forensic method for identification of confiscated illegal wildlife items. Separations 2020, 7, 5. [Google Scholar] [CrossRef]
- Van Steendam, K.; De Ceuleneer, M.; Dhaenens, M.; Van Hoofstat, D.; Deforce, D. Mass spectrometry-based proteomics as a tool to identify biological matrices in forensic science. Int. J. Leg. Med. 2013, 127, 287–298. [Google Scholar] [CrossRef]
- Tridico, S.R.; Houck, M.M.; Kirkbride, K.P.; Smith, M.E.; Yates, B.C. Morphological identification of animal hairs: Myths and misconceptions, possibilities and pitfalls. Forensic Sci. Int. 2014, 238, 101–107. [Google Scholar] [CrossRef]
- Kashyap, D.; Yadav, M.; Rathore, S.S.; Gupta, P.; Rane, P.; Uikey, B.N.; Kumar, C.; Chandrakar, T.R.; Jadav, K.; Jain, R. A Comparative Microscopic and Micrometric Analysis of Birds of Different Feather Types for Identification of Species. J. Int. Wildl. Law Policy 2024, 27, 235–246. [Google Scholar] [CrossRef]
- Trail, P.W. Identifying Bald versus Golden Eagle bones: A primer for wildlife biologists and law enforcement officers. J. Fish Wildl. Manag. 2017, 8, 596–610. [Google Scholar] [CrossRef]
- Garvin, H.M.; Dunn, R.; Sholts, S.B.; Litten, M.S.; Mohamed, M.; Kuttickat, N.; Skantz, N. Forensic tools for species identification of skeletal remains: Metrics, statistics, and OsteoID. Biology 2021, 11, 25. [Google Scholar] [CrossRef]
- Kufnerová, J.; Frouzová, J.; Světlík, I.; Weissová, K.; Brabcová, K.P. Expert opinion versus radiocarbon dating in the ivory trade. Glob. Ecol. Conserv. 2025, 61, e03659. [Google Scholar] [CrossRef]
- Prigge, T.L.; Andersson, A.A.; Hatten, C.E.; Leung, E.Y.; Baker, D.M.; Bonebrake, T.C.; Dingle, C. Wildlife trade investigations benefit from multivariate stable isotope analyses. Biol. Rev. 2025, 100, 1083–1104. [Google Scholar] [CrossRef] [PubMed]
- Brabcová, K.P.; Pravdíková, N.; Čápová, K.; Frouzová, J.; Hebenstreitová, K.; Jandová, K.; Kukla, J.; Rajmonová, E.; Salaba, O.; Světlík, I. Effect of leather tanning process on stable isotopes and radiocarbon in tissues of Persian leopard: Preliminary results. Forensic Sci. Int. Rep. 2024, 10, 100398. [Google Scholar] [CrossRef]
- Kufnerova, J. A novel approach of using shed skins of the green tree python, Morelia viridis, for forensic purposes. Eur. J. Environ. Sci. 2021, 11, 107–112. [Google Scholar] [CrossRef]




| Discipline | Primary Objectives | Examples of Typical Samples |
|---|---|---|
| Human DNA Forensics | Individual identification of a person of interest, victim, or missing person; parentage and kinship analysis; genealogical leads in criminal cases. | Blood, saliva, semen, hair with follicle, tissue, hair shaft, skin cells (touch DNA), bone, teeth. |
| Animal DNA Forensics | Species identification; determination of geographic origin; individual identification (less common); parentage and kinship analysis; confirmation of captive-bred vs. wild status. | Blood, tissue, hair, bone, feces, processed animal products (meat, leather, horn, ivory), vomit, stomach contents. |
| Focus Area | A Septennium Review of Wildlife Forensic DNA Analysis in South Africa | General Application Perspective |
|---|---|---|
| Tracking Judicial Impact | Unique recommendation: Track successes and challenges when forensic case reports are submitted to courts to understand the impact of genetic evidence on convictions and sentences, which is essential for obtaining further funding. | This aspect is not addressed, although it should be supported. Focus instead is on the scientific and technical steps required for maximizing typing success, and thus, before court submission. |
| Addressing Ambiguous Taxonomy | Highlights challenges with changing or unclear taxonomy for specific South African species (e.g., African elephants, giraffes) and the need for validated technologies that reflect these changes for forensic distinction. | Discusses the issue of hybridization and NUMTs. Does not focus on specific taxonomic reclassifications or legislative lags for endemic species as a primary challenge. A good recommendation for those working on specific species and distinguishing between close species. |
| Contamination/Lab Protocols | Focuses generally on training to avoid contamination. | Recommendations for laboratory protocols: Develop and validate DNA extraction and inhibitor removal protocols specifically for non-human samples, employ qPCR with an internal amplification control to accurately measure target DNA and detect inhibitors, discuss the use of heat-labile dsDNase to mitigate non-human contamination in reagents, and consider contaminating-human DNA as a potential investigative lead. |
| Standardization and Resources | Mentions following ISFG standards and addressing chronic underfunding. | Recommendations for standardization: Develop kits specifically for non-human identification, noting that ISO 18385 does not cover non-human DNA contaminants; suggest alternatives for obtaining CITES samples (collaborative studies, mimicked DNA, Memoranda of Understanding with zoos) if shipping restrictions cannot be overcome. Also notes under funding issues. |
| Multidisciplinary Approach | Not a primary recommendation, but necessity is implied by the technical hurdles. | Recommendation to adopt a multidisciplinary approach, utilizing methods like profiling of volatilomes, mass spectrometry, hair morphology, osteology, and radiocarbon dating as additional or orthogonal tests. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budowle, B.; Sajantila, A.; Vanek, D. Animal Species and Identity Testing: Developments, Challenges, and Applications to Non-Human Forensics. Genes 2025, 16, 1503. https://doi.org/10.3390/genes16121503
Budowle B, Sajantila A, Vanek D. Animal Species and Identity Testing: Developments, Challenges, and Applications to Non-Human Forensics. Genes. 2025; 16(12):1503. https://doi.org/10.3390/genes16121503
Chicago/Turabian StyleBudowle, Bruce, Antti Sajantila, and Daniel Vanek. 2025. "Animal Species and Identity Testing: Developments, Challenges, and Applications to Non-Human Forensics" Genes 16, no. 12: 1503. https://doi.org/10.3390/genes16121503
APA StyleBudowle, B., Sajantila, A., & Vanek, D. (2025). Animal Species and Identity Testing: Developments, Challenges, and Applications to Non-Human Forensics. Genes, 16(12), 1503. https://doi.org/10.3390/genes16121503








