Addressing Ancestral Underrepresentation in Oncobiology: The Need for Sub-Saharan African-Specific In Vitro Models
Abstract
1. Introduction
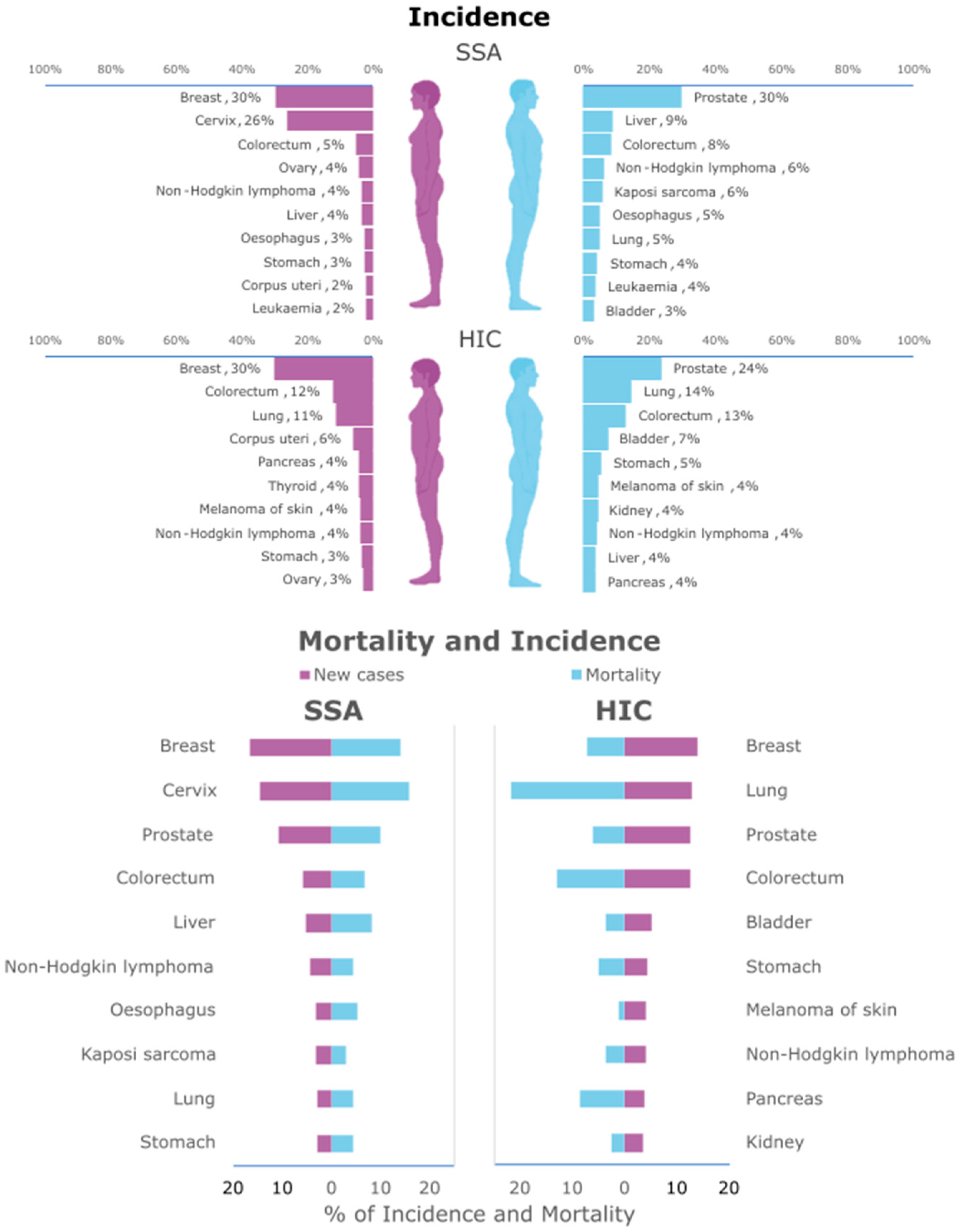
2. Cancer in SSA
3. Cancer Genomics in People of SSA Ancestry
4. In Vitro Models
4.1. Cancer Cell Lines (CCLs)
4.1.1. Historical Contextualization
4.1.2. Cancer Cell Line Repositories
4.1.3. Omics Characterization of Cancer Cell Line Panels
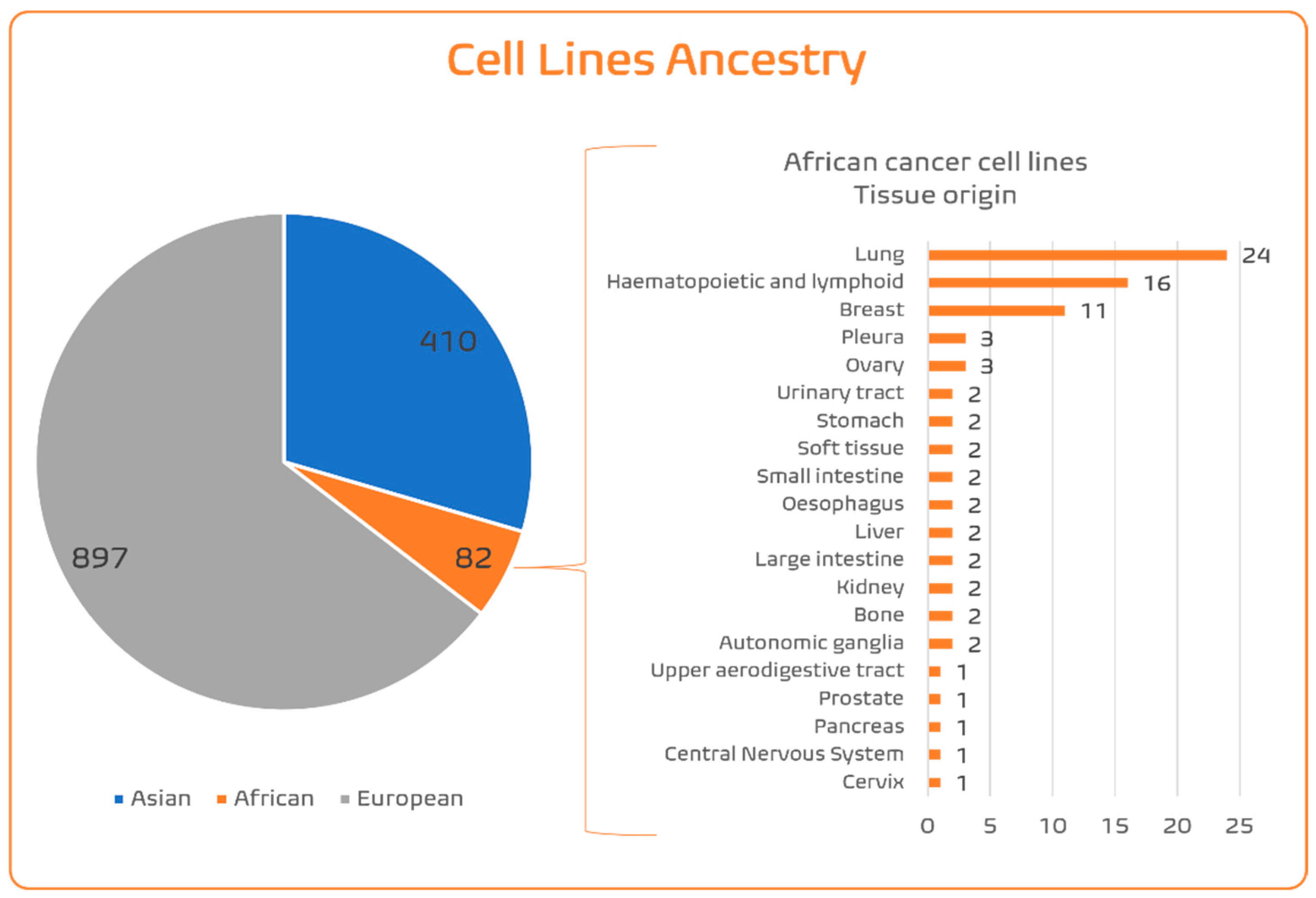
4.1.4. SSA Ancestry Representativeness in CCL Panels
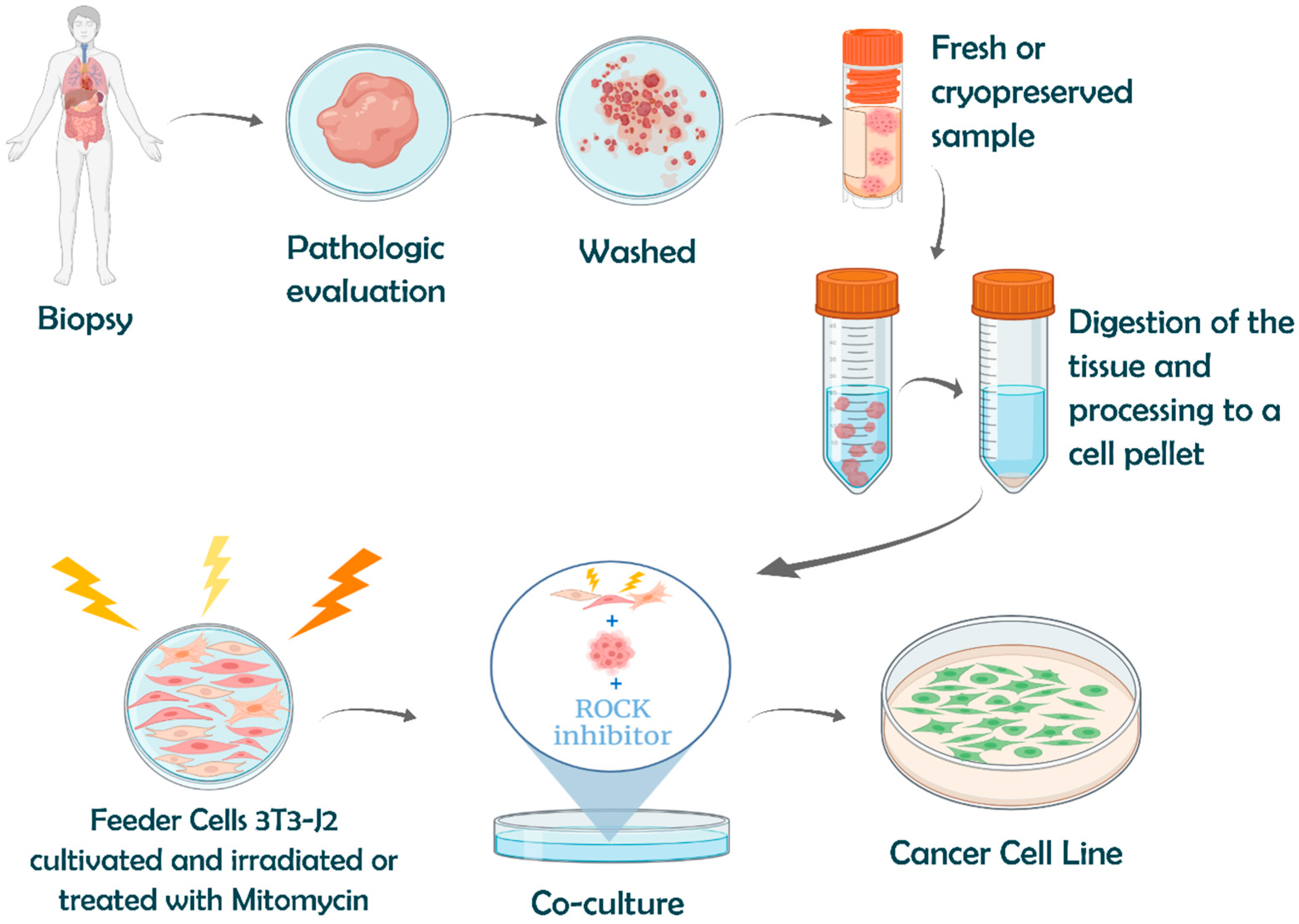
4.1.5. Establishment of Cancer Cell Lines: Surpassing Normal Cell Death
Conventional Methods of Cell Immortalization
Advanced Immortalization Techniques: Conditional Reprogramming
4.2. Advances in Pluripotency and Three-Dimensional Modelling
4.2.1. Human-Induced Pluripotent Stem Cells (hiPSCs)
4.2.2. Organoids
5. Potential for Cancer Translation in SSA Patients
6. Implementing Oncobiology Studies with Cell Modelling Derived from SSA Patients
Case Study: PALOP
- Source of cancer patient tissue: Fresh cancer tissues from PALOP or diaspora patients are to be collected from surgeries and biopsies conducted in hospitals from the Portuguese Health System (SNS, in the Portuguese abbreviation). The PALOP diaspora community residing in Portugal and its descendants have been increasing since 1975, and especially so since the year 2017. Additionally, the SNS has agreements with PALOP health systems for transfer to Portugal and local treatment (including surgeries) of PALOP resident cancer patients. This has the additional advantage of all PALOP countries being represented in the panel, as this diaspora community is multi-ethnic.
- Establishment of PALOP advanced cell models: Collecting these samples in Portugal allows us the opportunity to establish PALOP advanced cell models in well-equipped Portuguese laboratories while avoiding the difficulties associated with transporting live cells across countries and continents. The CR technique can be used to establish the PALOP CCLs. First efforts should prioritize underrepresented tissues in the few SSA CCLs available in international panels, such as cervix, pancreas, prostate, and kidney tissues. Our very preliminary results indicate that changes to the standard CR protocol [99] may be needed to avoid bacterial contamination (from the sample) in tissues with microbiota (in particular, tissues from the digestive and female reproductive systems).
- Capacitating PALOP medical doctors, researchers and technicians: Of greatest importance is the training of PALOP medical doctors, researchers, and technicians. There are many protocols established in Portuguese hospitals for the training of PALOP surgeons and oncologists. These visiting PALOP MDs must be involved in the collection of the material for the establishment of PALOP CCLs. Specific training must be established in Portuguese research institutions to train PALOP researchers and technicians in cell culturing techniques and in conducting in vitro experiments. Funding for this training is more easily obtained through PhD grants, which also offer the added benefit of extended (usually four-year) training and the development of an independent researcher.
- Funding of Portuguese–PALOP partnership: Through a strong network of North–South research partnerships, PALOP scientists can leapfrog technological gaps and build a research ecosystem capable of conducting and leading advanced oncobiology research. This strategy will also allow the PALOP research community to affirm itself within the African continent, on par with English- and French-speaking communities, for instance in the African Organisation for Research and Training in Cancer (AORTIC; https://aortic-africa.org/; accessed on 17 November 2025) and other Health Hubs.
7. Conclusions and Future Perspectives
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AA | African American |
| ASC | Adult Stem Cell |
| ATCC | American Type Culture Collection |
| BC | Breast Cancer |
| CCL | Cancer Cell Line |
| CCLE | Cancer Cell Line Encyclopedia |
| CR | Conditional Reprogramming |
| ECACC | European Collection of Authenticated Cell Cultures |
| GDSC | Genomics of Drug Sensitivity in Cancer |
| GWAS | Genome-Wide Association Study |
| HCMI | Human Cancer Models Initiative |
| HIC | High-Income Country |
| HPV | Human Papillomavirus |
| JCRB | Japanese Cancer Research Resources Bank |
| KCLB | Korean Cell Line Bank |
| LIC | Low-Income Country |
| PALOP | Portuguese-Speaking African Country |
| PDO | Patients-Derived Organoid |
| PDX | Patient-Derived Xenograph |
| SNP | Single-Nucleotide Polymorphism |
| SSA | Sub-Saharan Africa |
| STR | Short Tandem Repeat |
| TCGA | The Cancer Genome Atlas Program |
| TNBC | Triple-Negative Breast Cancer |
| USA | United States America |
References
- Pereira, L.; Mutesa, L.; Tindana, P.; Ramsay, M. African genetic diversity and adaptation inform a precision medicine agenda. Nat. Rev. Genet. 2021, 22, 284–306. [Google Scholar] [CrossRef]
- Quintana-Murci, L. Human Immunology through the Lens of Evolutionary Genetics. Cell 2019, 177, 184–199. [Google Scholar] [CrossRef]
- Grossman, S.R.; Andersen, K.G.; Shlyakhter, I.; Tabrizi, S.; Winnicki, S.; Yen, A.; Park, D.J.; Griesemer, D.; Karlsson, E.K.; Wong, S.H.; et al. Identifying recent adaptations in large-scale genomic data. Cell 2013, 152, 703–713. [Google Scholar] [CrossRef]
- Pedro, N.; Pinto, R.J.; Cavadas, B.; Pereira, L. Sub-Saharan African information potential to unveil adaptations to infectious disease. Hum. Mol. Genet. 2021, 30, R138–R145. [Google Scholar] [CrossRef]
- Ngwa, W.; Addai, B.W.; Adewole, I.; Ainsworth, V.; Alaro, J.; Alatise, O.I.; Ali, Z.; Anderson, B.O.; Anorlu, R.; Avery, S.; et al. Cancer in sub-Saharan Africa: A Lancet Oncology Commission. Lancet Oncol. 2022, 23, e251–e312. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Gurdasani, D.; Carstensen, T.; Fatumo, S.; Chen, G.; Franklin, C.S.; Prado-Martinez, J.; Bouman, H.; Abascal, F.; Haber, M.; Tachmazidou, I.; et al. Uganda Genome Resource Enables Insights into Population History and Genomic Discovery in Africa. Cell 2019, 179, 984–1002.e1036. [Google Scholar] [CrossRef]
- Gurdasani, D.; Barroso, I.; Zeggini, E.; Sandhu, M.S. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet. 2019, 20, 520–535. [Google Scholar] [CrossRef]
- Spratt, D.E.; Chan, T.; Waldron, L.; Speers, C.; Feng, F.Y.; Ogunwobi, O.O.; Osborne, J.R. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol. 2016, 2, 1070–1074. [Google Scholar] [CrossRef]
- Park, S.L.; Cheng, I.; Haiman, C.A. Genome-Wide Association Studies of Cancer in Diverse Populations. Cancer Epidemiol. Biomark. Prev. 2018, 27, 405–417. [Google Scholar] [CrossRef]
- Rebbeck, T.R. Cancer in sub-Saharan Africa. Science 2020, 367, 27–28. [Google Scholar] [CrossRef]
- Rowe, M.; Fitzsimmons, L.; Bell, A.I. Epstein-Barr virus and Burkitt lymphoma. Chin. J. Cancer 2014, 33, 609–619. [Google Scholar] [CrossRef]
- Parkin, D.M.; Hämmerl, L.; Ferlay, J.; Kantelhardt, E.J. Cancer in Africa 2018: The role of infections. Int. J. Cancer 2020, 146, 2089–2103. [Google Scholar] [CrossRef]
- Chinula, L.; Moses, A.; Gopal, S. HIV-associated malignancies in sub-Saharan Africa: Progress, challenges, and opportunities. Curr. Opin. HIV AIDS 2017, 12, 89–95. [Google Scholar] [CrossRef]
- Gurdasani, D.; Carstensen, T.; Tekola-Ayele, F.; Pagani, L.; Tachmazidou, I.; Hatzikotoulas, K.; Karthikeyan, S.; Iles, L.; Pollard, M.O.; Choudhury, A.; et al. The African Genome Variation Project shapes medical genetics in Africa. Nature 2015, 517, 327–332. [Google Scholar] [CrossRef]
- Patin, E.; Lopez, M.; Grollemund, R.; Verdu, P.; Harmant, C.; Quach, H.; Laval, G.; Perry, G.H.; Barreiro, L.B.; Froment, A.; et al. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science 2017, 356, 543–546. [Google Scholar] [CrossRef]
- Triska, P.; Soares, P.; Patin, E.; Fernandes, V.; Cerny, V.; Pereira, L. Extensive Admixture and Selective Pressure Across the Sahel Belt. Genome Biol. Evol. 2015, 7, 3484–3495. [Google Scholar] [CrossRef]
- Brucato, N.; Fernandes, V.; Mazieres, S.; Kusuma, P.; Cox, M.P.; Ng’ang’a, J.W.; Omar, M.; Simeone-Senelle, M.C.; Frassati, C.; Alshamali, F.; et al. The Comoros Show the Earliest Austronesian Gene Flow into the Swahili Corridor. Am. J. Hum. Genet. 2018, 102, 58–68. [Google Scholar] [CrossRef]
- Lin, J.; Musunuru, K. From Genotype to Phenotype: A Primer on the Functional Follow-up of Genome-Wide Association Studies in Cardiovascular Disease. Circ. Genom. Precis. Med. 2018, 11, e001946. [Google Scholar]
- Cavadas, B.; Leite, M.; Pedro, N.; Magalhães, A.C.; Melo, J.; Correia, M.; Máximo, V.; Camacho, R.; Fonseca, N.A.; Figueiredo, C.; et al. Shedding Light on the African Enigma: In Vitro Testing of Homo sapiens-Helicobacter pylori Coevolution. Microorganisms 2021, 9, 240. [Google Scholar] [CrossRef]
- Parkin, D.M.; Ferlay, J.; Jemal, A.; Borok, M.; Manraj, S.S.; N’da, G.G.; Ogunbiyi, J.O.; Liu, B.; Bray, F. Cancer in Sub-Saharan Africa; IARC Scientific Publications: Lyon, France, 2018; Volume 167. [Google Scholar]
- Hamdi, Y.; Abdeljaoued-Tej, I.; Zatchi, A.A.; Abdelhak, S.; Boubaker, S.; Brown, J.S.; Benkahla, A. Cancer in Africa: The Untold Story. Front. Oncol. 2021, 11, 650117. [Google Scholar] [CrossRef]
- Miguel, F.; Lopes, L.V.; Ferreira, E.; Ribas, E.; Pelaez, A.F.; Leal, C.; Amaro, T.; Lopes, P.; Santos, C.M.; Lopes, C.; et al. Breast cancer in Angola, molecular subtypes: A first glance. Ecancermedicalscience 2017, 11, 763. [Google Scholar] [CrossRef]
- Brandão, M.; Guisseve, A.; Bata, G.; Alberto, M.; Ferro, J.; Garcia, C.; Zaqueu, C.; Lorenzoni, C.; Leitão, D.; Come, J.; et al. Breast cancer subtypes: Implications for the treatment and survival of patients in Africa-a prospective cohort study from Mozambique. ESMO Open 2020, 5, e000829. [Google Scholar] [CrossRef]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Tse, C.K.; Edmiston, S.; et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. BCR 2020, 22, 61. [Google Scholar] [CrossRef]
- Come, J.; Pereira, J.B.; Pinto, R.; Carrilho, C.; Pereira, L.; Lara Santos, L. The Upper Digestive Tract Microbiome and Oesophageal Squamous Cell Carcinoma: Epidemiology, Pathogenesis, and Clinical Implications in Africa. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2021, 88, 141–155. [Google Scholar] [CrossRef]
- Carrilho, C.; Modcoicar, P.; Cunha, L.; Ismail, M.; Guisseve, A.; Lorenzoni, C.; Fernandes, F.; Peleteiro, B.; Almeida, R.; Figueiredo, C.; et al. Prevalence of Helicobacter pylori infection, chronic gastritis, and intestinal metaplasia in Mozambican dyspeptic patients. Virchows Arch. 2009, 454, 153–160. [Google Scholar] [CrossRef]
- Lopes, L.V.; Conceição, A.V.; Oliveira, J.B.; Tavares, A.; Domingos, C.; Santos, L.L. Cancer in Angola, resources and strategy for its control. Pan Afr. Med. J. 2012, 12, 13. [Google Scholar]
- Kodaman, N.; Pazos, A.; Schneider, B.G.; Piazuelo, M.B.; Mera, R.; Sobota, R.S.; Sicinschi, L.A.; Shaffer, C.L.; Romero-Gallo, J.; de Sablet, T.; et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc. Natl. Acad. Sci. USA 2014, 111, 1455–1460. [Google Scholar] [CrossRef]
- Kramer, J. Eradicating cervical cancer: Lessons learned from Rwanda and Australia. Int. J. Gynaecol. Obstet. 2021, 154, 270–276. [Google Scholar] [CrossRef]
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Gunes, O.; Hall, P.; Hayhurst, J.; et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic Acids Res. 2023, 51, D977–D985. [Google Scholar] [CrossRef]
- Abdellaoui, A.; Yengo, L.; Verweij, K.J.H.; Visscher, P.M. 15 years of GWAS discovery: Realizing the promise. Am. J. Hum. Genet. 2023, 110, 179–194. [Google Scholar] [CrossRef]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Duncan, L.; Shen, H.; Gelaye, B.; Meijsen, J.; Ressler, K.; Feldman, M.; Peterson, R.; Domingue, B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019, 10, 3328. [Google Scholar] [CrossRef]
- Lee, H.; Palm, J.; Grimes, S.M.; Ji, H.P. The Cancer Genome Atlas Clinical Explorer: A web and mobile interface for identifying clinical-genomic driver associations. Genome Med. 2015, 7, 112. [Google Scholar] [CrossRef]
- Carrot-Zhang, J.; Chambwe, N.; Damrauer, J.S.; Knijnenburg, T.A.; Robertson, A.G.; Yau, C.; Zhou, W.; Berger, A.C.; Huang, K.L.; Newberg, J.Y.; et al. Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer. Cancer Cell 2020, 37, 639–654.e636. [Google Scholar] [CrossRef]
- Mori, J.O.; White, J.; Elhussin, I.; Duduyemi, B.M.; Karanam, B.; Yates, C.; Wang, H. Molecular and pathological subtypes related to prostate cancer disparities and disease outcomes in African American and European American patients. Front. Oncol. 2022, 12, 928357. [Google Scholar] [CrossRef]
- Krishnan, B.; Rose, T.L.; Kardos, J.; Milowsky, M.I.; Kim, W.Y. Intrinsic Genomic Differences Between African American and White Patients with Clear Cell Renal Cell Carcinoma. JAMA Oncol. 2016, 2, 664–667. [Google Scholar] [CrossRef]
- Pinto, R.J.; Ferreira, D.; Salamanca, P.; Miguel, F.; Borges, P.; Barbosa, C.; Costa, V.; Lopes, C.; Santos, L.L.; Pereira, L. Coding and regulatory somatic profiling of triple-negative breast cancer in Sub-Saharan African patients. Sci. Rep. 2025, 15, 10325. [Google Scholar] [CrossRef]
- Pitt, J.J.; Riester, M.; Zheng, Y.; Yoshimatsu, T.F.; Sanni, A.; Oluwasola, O.; Veloso, A.; Labrot, E.; Wang, S.; Odetunde, A.; et al. Characterization of Nigerian breast cancer reveals prevalent homologous recombination deficiency and aggressive molecular features. Nat. Commun. 2018, 9, 4181. [Google Scholar] [CrossRef]
- Saleh, M.; Chandrashekar, D.S.; Shahin, S.; Agarwal, S.; Kim, H.G.; Behring, M.; Shaikh, A.J.; Moloo, Z.; Eltoum, I.A.; Yates, C.; et al. Comparative analysis of triple-negative breast cancer transcriptomics of Kenyan, African American and Caucasian Women. Transl. Oncol. 2021, 14, 101086. [Google Scholar] [CrossRef]
- Martini, R.; Delpe, P.; Chu, T.R.; Arora, K.; Lord, B.; Verma, A.; Bedi, D.; Karanam, B.; Elhussin, I.; Chen, Y.; et al. African Ancestry-Associated Gene Expression Profiles in Triple-Negative Breast Cancer Underlie Altered Tumor Biology and Clinical Outcome in Women of African Descent. Cancer Discov. 2022, 12, 2530–2551. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; Teo, S.H.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018, 39, 593–620. [Google Scholar] [CrossRef]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Gillet, J.P.; Varma, S.; Gottesman, M.M. The clinical relevance of cancer cell lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef]
- Geraghty, R.J.; Capes-Davis, A.; Davis, J.M.; Downward, J.; Freshney, R.I.; Knezevic, I.; Lovell-Badge, R.; Masters, J.R.; Meredith, J.; Stacey, G.N.; et al. Guidelines for the use of cell lines in biomedical research. Br. J. Cancer 2014, 111, 1021–1046. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Drexler, H.G. Guidelines for the characterization and publication of human malignant hematopoietic cell lines. Leukemia 1999, 13, 835–842. [Google Scholar] [CrossRef]
- Matsuo, Y. Establishment and characterization of human B cell precursor-leukemia cell lines. Leuk. Res. 1998, 22, 567–579. [Google Scholar] [CrossRef]
- Masters, J.R. HeLa cells 50 years on: The good, the bad and the ugly. Nat. Rev. Cancer 2002, 2, 315–319. [Google Scholar] [CrossRef]
- McCullough, D.C. Metamorphosis and Resurrection of Henrietta Lacks. Cult. Stud. Crit. Methodol. 2020, 20, 251–260. [Google Scholar] [CrossRef]
- Sodeke, S.O.; Powell, L.R. Paying Tribute to Henrietta Lacks at Tuskegee University and at The Virginia Henrietta Lacks Commission, Richmond, Virginia. J. Health Care Poor Underserved 2019, 30, 1–11. [Google Scholar] [CrossRef]
- Pulvertaft, R.J.V. Cytology Of Burkitt’s Tumour (African Lymphoma). Lancet 1964, 283, 238–240. [Google Scholar] [CrossRef]
- Gartler, S.M. Apparent Hela cell contamination of human heteroploid cell lines. Nature 1968, 217, 750–751. [Google Scholar] [CrossRef]
- Capes-Davis, A.; Theodosopoulos, G.; Atkin, I.; Drexler, H.G.; Kohara, A.; MacLeod, R.A.; Masters, J.R.; Nakamura, Y.; Reid, Y.A.; Reddel, R.R.; et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer 2010, 127, 1–8. [Google Scholar] [CrossRef]
- Coecke, S.; Balls, M.; Bowe, G.; Davis, J.; Gstraunthaler, G.; Hartung, T.; Hay, R.; Merten, O.W.; Price, A.; Schechtman, L.; et al. Guidance on good cell culture practice. a report of the second ECVAM task force on good cell culture practice. Altern. Lab. Anim. 2005, 33, 261–287. [Google Scholar] [CrossRef]
- Ling, A.; Gruener, R.F.; Fessler, J.; Huang, R.S. More than fishing for a cure: The promises and pitfalls of high throughput cancer cell line screens. Pharmacol. Ther. 2018, 191, 178–189. [Google Scholar] [CrossRef]
- Kim, H.S.; Sung, Y.J.; Paik, S. Cancer Cell Line Panels Empower Genomics-Based Discovery of Precision Cancer Medicine. Yonsei Med. J. 2015, 56, 1186–1198. [Google Scholar] [CrossRef]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Workman, P. The NCI-60 Human Tumor Cell Line Screen: A Catalyst for Progressive Evolution of Models for Discovery and Development of Cancer Drugs. Cancer Res. 2023, 83, 3170–3173. [Google Scholar] [CrossRef]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–122. [Google Scholar] [CrossRef]
- Adams, S.; Robbins, F.M.; Chen, D.; Wagage, D.; Holbeck, S.L.; Morse, H.C., 3rd; Stroncek, D.; Marincola, F.M. HLA class I and II genotype of the NCI-60 cell lines. J. Transl. Med. 2005, 3, 11. [Google Scholar] [CrossRef]
- Roschke, A.V.; Tonon, G.; Gehlhaus, K.S.; McTyre, N.; Bussey, K.J.; Lababidi, S.; Scudiero, D.A.; Weinstein, J.N.; Kirsch, I.R. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003, 63, 8634–8647. [Google Scholar]
- Lorenzi, P.L.; Reinhold, W.C.; Varma, S.; Hutchinson, A.A.; Pommier, Y.; Chanock, S.J.; Weinstein, J.N. DNA fingerprinting of the NCI-60 cell line panel. Mol. Cancer Ther. 2009, 8, 713–724. [Google Scholar] [CrossRef]
- Kong, D.; Yamori, T. JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs. Bioorg. Med. Chem. 2012, 20, 1947–1951. [Google Scholar] [CrossRef]
- Yamori, T. Panel of human cancer cell lines provides valuable database for drug discovery and bioinformatics. Cancer Chemother. Pharmacol. 2003, 52 (Suppl. S1), S74–S79. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Jaffe, J.D.; Wang, Y.; Chan, H.M.; Zhang, J.; Huether, R.; Kryukov, G.V.; Bhang, H.E.; Taylor, J.E.; Hu, M.; Englund, N.P.; et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 1386–1391. [Google Scholar] [CrossRef]
- Li, H.; Ning, S.; Ghandi, M.; Kryukov, G.V.; Gopal, S.; Deik, A.; Souza, A.; Pierce, K.; Keskula, P.; Hernandez, D.; et al. The landscape of cancer cell line metabolism. Nat. Med. 2019, 25, 850–860. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R., 3rd; Kalocsay, M.; Jane-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e316. [Google Scholar] [CrossRef]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Gonçalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e516. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Goncalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Lane-Reticker, S.K.; Manguso, R.T.; Haining, W.N. Pooled in vivo screens for cancer immunotherapy target discovery. Immunotherapy 2018, 10, 167–170. [Google Scholar] [CrossRef]
- Bryc, K.; Durand, E.Y.; Macpherson, J.M.; Reich, D.; Mountain, J.L. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015, 96, 37–53. [Google Scholar] [CrossRef]
- Dutil, J.; Chen, Z.; Monteiro, A.N.; Teer, J.K.; Eschrich, S.A. An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res. 2019, 79, 1263–1273. [Google Scholar]
- Genomes Project, C.; Abecasis, G.R.; Altshuler, D.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Gibbs, R.A.; Hurles, M.E.; McVean, G.A. A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar]
- Chapman, S.; Liu, X.; Meyers, C.; Schlegel, R.; McBride, A.A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Investig. 2010, 120, 2619–2626. [Google Scholar] [CrossRef]
- Boehm, J.S.; Hahn, W.C. Immortalized cells as experimental models to study cancer. Cytotechnology 2004, 45, 47–59. [Google Scholar]
- Wei, W.; Sedivy, J.M. Differentiation between senescence (M1) and crisis (M2) in human fibroblast cultures. Exp. Cell Res. 1999, 253, 519–522. [Google Scholar]
- Stewart, S.A.; Weinberg, R.A. Senescence: Does it all happen at the ends? Oncogene 2002, 21, 627–630. [Google Scholar]
- Falandry, C.; Bonnefoy, M.; Freyer, G.; Gilson, E. Biology of Cancer and Aging: A Complex Association with Cellular Senescence. J. Clin. Oncol. 2014, 32, 2604–2610. [Google Scholar]
- Hawley-Nelson, P.; Vousden, K.H.; Hubbert, N.L.; Lowy, D.R.; Schiller, J.T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989, 8, 3905–3910. [Google Scholar] [CrossRef]
- Van der Haegen, B.A.; Shay, J.W. Immortalization of human mammary epithelial cells by SV40 large T-antigen involves a two step mechanism. Vitr. Cell. Dev. Biol. 1993, 29A Pt 1, 180–182. [Google Scholar] [CrossRef]
- Ramboer, E.; De Craene, B.; De Kock, J.; Vanhaecke, T.; Berx, G.; Rogiers, V.; Vinken, M. Strategies for immortalization of primary hepatocytes. J. Hepatol. 2014, 61, 925–943. [Google Scholar] [CrossRef]
- Liu, X.; Dakic, A.; Chen, R.; Disbrow, G.L.; Zhang, Y.; Dai, Y.; Schlegel, R. Cell-restricted immortalization by human papillomavirus correlates with telomerase activation and engagement of the hTERT promoter by Myc. J. Virol. 2008, 82, 11568–11576. [Google Scholar] [CrossRef]
- Charette, S.T.; McCance, D.J. The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene 2007, 26, 7386–7390. [Google Scholar] [CrossRef]
- Trakarnsanga, K.; Griffiths, R.E.; Wilson, M.C.; Blair, A.; Satchwell, T.J.; Meinders, M.; Cogan, N.; Kupzig, S.; Kurita, R.; Nakamura, Y.; et al. An immortalized adult human erythroid line facilitates sustainable and scalable generation of functional red cells. Nat. Commun. 2017, 8, 14750. [Google Scholar] [CrossRef]
- Shin, H.Y.; Yang, W.; Lee, E.J.; Han, G.H.; Cho, H.; Chay, D.B.; Kim, J.H. Establishment of five immortalized human ovarian surface epithelial cell lines via SV40 T antigen or HPV E6/E7 expression. PLoS ONE 2018, 13, e0205297. [Google Scholar] [CrossRef]
- Hung, C.L.; Maiuri, T.; Bowie, L.E.; Gotesman, R.; Son, S.; Falcone, M.; Giordano, J.V.; Gillis, T.; Mattis, V.; Lau, T.; et al. A patient-derived cellular model for Huntington’s disease reveals phenotypes at clinically relevant CAG lengths. Mol. Biol. Cell 2018, 29, 2809–2820. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef]
- Roig, A.I.; Eskiocak, U.; Hight, S.K.; Kim, S.B.; Delgado, O.; Souza, R.F.; Spechler, S.J.; Wright, W.E.; Shay, J.W. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology 2010, 138, 1012–1021.e5. [Google Scholar] [CrossRef]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Liu, X.; Krawczyk, E.; Suprynowicz, F.A.; Palechor-Ceron, N.; Yuan, H.; Dakic, A.; Simic, V.; Zheng, Y.L.; Sripadhan, P.; Chen, C.; et al. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 2017, 12, 439–451. [Google Scholar] [CrossRef]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef]
- Suprynowicz, F.A.; Upadhyay, G.; Krawczyk, E.; Kramer, S.C.; Hebert, J.D.; Liu, X.; Yuan, H.; Cheluvaraju, C.; Clapp, P.W.; Boucher, R.C., Jr.; et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20035–20040. [Google Scholar] [CrossRef]
- Wu, X.; Wang, S.; Li, M.; Li, J.; Shen, J.; Zhao, Y.; Pang, J.; Wen, Q.; Chen, M.; Wei, B.; et al. Conditional reprogramming: Next generation cell culture. Acta Pharm. Sin. B 2020, 10, 1360–1381. [Google Scholar] [CrossRef]
- Dakic, A.; DiVito, K.; Fang, S.; Suprynowicz, F.; Gaur, A.; Li, X.; Palechor-Ceron, N.; Simic, V.; Choudhury, S.; Yu, S.; et al. ROCK inhibitor reduces Myc-induced apoptosis and mediates immortalization of human keratinocytes. Oncotarget 2016, 7, 66740–66753. [Google Scholar] [CrossRef]
- Mondal, A.M.; Zhou, H.; Horikawa, I.; Suprynowicz, F.A.; Li, G.; Dakic, A.; Rosenthal, B.; Ye, L.; Harris, C.C.; Schlegel, R.; et al. Δ133p53α, a natural p53 isoform, contributes to conditional reprogramming and long-term proliferation of primary epithelial cells. Cell Death Dis. 2018, 9, 750. [Google Scholar] [CrossRef]
- Santos, A.; Bakker, A.D.; de Blieck-Hogervorst, J.M.; Klein-Nulend, J. WNT5A induces osteogenic differentiation of human adipose stem cells via rho-associated kinase ROCK. Cytotherapy 2010, 12, 924–932. [Google Scholar] [CrossRef]
- Ji, H.; Tang, H.; Lin, H.; Mao, J.; Gao, L.; Liu, J.; Wu, T. Rho/Rock cross-talks with transforming growth factor-β/Smad pathway participates in lung fibroblast-myofibroblast differentiation. Biomed. Rep. 2014, 2, 787–792. [Google Scholar] [CrossRef]
- Saenz, F.R.; Ory, V.; AlOtaiby, M.; Rosenfield, S.; Furlong, M.; Cavalli, L.R.; Johnson, M.D.; Liu, X.; Schlegel, R.; Wellstein, A.; et al. Conditionally reprogrammed normal and transformed mouse mammary epithelial cells display a progenitor-cell-like phenotype. PLoS ONE 2014, 9, e97666. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, J.; Yang, C.; Tan, R.; Hou, J.; Shi, Y.; Zhang, H.; Ma, S.; Wang, J.; Zhang, M.; et al. Continuous culture of urine-derived bladder cancer cells for precision medicine. Protein Cell 2019, 10, 902–907. [Google Scholar] [CrossRef]
- Kodack, D.P.; Farago, A.F.; Dastur, A.; Held, M.A.; Dardaei, L.; Friboulet, L.; von Flotow, F.; Damon, L.J.; Lee, D.; Parks, M.; et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep. 2017, 21, 3298–3309. [Google Scholar] [CrossRef]
- Mahajan, A.S.; Sugita, B.M.; Duttargi, A.N.; Saenz, F.; Krawczyk, E.; McCutcheon, J.N.; Fonseca, A.S.; Kallakury, B.; Pohlmann, P.; Gusev, Y.; et al. Genomic comparison of early-passage conditionally reprogrammed breast cancer cells to their corresponding primary tumors. PLoS ONE 2017, 12, e0186190. [Google Scholar] [CrossRef]
- Park, G.; Rim, Y.A.; Sohn, Y.; Nam, Y.; Ju, J.H. Replacing Animal Testing with Stem Cell-Organoids: Advantages and Limitations. Stem Cell Rev. Rep. 2024, 20, 1375–1386. [Google Scholar] [CrossRef]
- Prochazkova, M.; Chavez, M.G.; Prochazka, J.; Felfy, H.; Mushegyan, V.; Klein, O.D. Chapter 18—Embryonic Versus Adult Stem Cells. In Stem Cell Biology and Tissue Engineering in Dental Sciences; Vishwakarma, A., Sharpe, P., Shi, S., Ramalingam, M., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 249–262. [Google Scholar]
- de Miguel-Beriain, I. The ethics of stem cells revisited. Adv. Drug Deliv. Rev. 2015, 82–83, 176–180. [Google Scholar] [CrossRef]
- Lo, B.; Parham, L. Ethical issues in stem cell research. Endocr. Rev. 2009, 30, 204–213. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, S.; Joo, J.Y.; Zhu, S.; Han, D.W.; Lin, T.; Trauger, S.; Bien, G.; Yao, S.; Zhu, Y.; et al. Generation of Induced Pluripotent Stem Cells Using Recombinant Proteins. Cell Stem Cell 2009, 4, 381–384. [Google Scholar]
- Kim, D.; Kim, C.H.; Moon, J.I.; Chung, Y.G.; Chang, M.Y.; Han, B.S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar]
- Shankaran, A.; Prasad, K.; Chaudhari, S.; Brand, A.; Satyamoorthy, K. Advances in development and application of human organoids. 3 Biotech 2021, 11, 257. [Google Scholar] [CrossRef]
- Adegunsoye, A.; Gonzales, N.M.; Gilad, Y. Induced Pluripotent Stem Cells in Disease Biology and the Evidence for Their In Vitro Utility. Annu. Rev. Genet. 2023, 57, 341–360. [Google Scholar] [CrossRef]
- Clark, B.J.; Lelos, M.J.; Loring, J.F. Advancing Parkinson’s disease treatment: Cell replacement therapy with neurons derived from pluripotent stem cells. Stem Cells 2024, 42, 781–790. [Google Scholar] [CrossRef]
- Scesa, G.; Adami, R.; Bottai, D. iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter? Cells 2021, 10, 1470. [Google Scholar] [CrossRef]
- Panopoulos, A.D.; D’Antonio, M.; Benaglio, P.; Williams, R.; Hashem, S.I.; Schuldt, B.M.; DeBoever, C.; Arias, A.D.; Garcia, M.; Nelson, B.C.; et al. iPSCORE: A Resource of 222 iPSC Lines Enabling Functional Characterization of Genetic Variation across a Variety of Cell Types. Stem Cell Rep. 2017, 8, 1086–1100. [Google Scholar] [CrossRef]
- Kilpinen, H.; Goncalves, A.; Leha, A.; Afzal, V.; Alasoo, K.; Ashford, S.; Bala, S.; Bensaddek, D.; Casale, F.P.; Culley, O.J.; et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 2017, 546, 370–375. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernandez-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Berkers, G.; van Mourik, P.; Vonk, A.M.; Kruisselbrink, E.; Dekkers, J.F.; de Winter-de Groot, K.M.; Arets, H.G.M.; Marck-van der Wilt, R.E.P.; Dijkema, J.S.; Vanderschuren, M.M.; et al. Rectal Organoids Enable Personalized Treatment of Cystic Fibrosis. Cell Rep. 2019, 26, 1701–1708.e1703. [Google Scholar] [CrossRef]
- Madorsky Rowdo, F.P.; Martini, R.; Ackermann, S.E.; Tang, C.P.; Tranquille, M.; Irizarry, A.; Us, I.; Alawa, O.; Moyer, J.E.; Sigouros, M.; et al. Kinome-Focused CRISPR-Cas9 Screens in African Ancestry Patient-Derived Breast Cancer Organoids Identify Essential Kinases and Synergy of EGFR and FGFR1 Inhibition. Cancer Res. 2025, 85, 551–566. [Google Scholar] [CrossRef]
- Usman, H.; Witonsky, D.; Bielski, M.C.; Lawrence, K.M.; Laxman, B.; Kupfer, S.S. Genomic and cellular responses to aspirin in colonic organoids from African- and European-Americans. Physiol. Genom. 2025, 57, 103–114. [Google Scholar] [CrossRef]
- Devall, M.; Eaton, S.; Yoshida, C.; Powell, S.M.; Casey, G.; Li, L. Assessment of Colorectal Cancer Risk Factors through the Application of Network-Based Approaches in a Racially Diverse Cohort of Colon Organoid Stem Cells. Cancers 2023, 15, 3550. [Google Scholar] [CrossRef]
- Abdulla, N.; Aronson, R.; Plessis, T.D.; Bebington, B.; Kaur, M. Protocol for the establishment and characterization of South African patient-derived intestinal organoids. STAR Protoc. 2025, 6, 103970. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Zhou, H.; Stoneking, M.; Tang, K. Global patterns of genetic diversity and signals of natural selection for human ADME genes. Hum. Mol. Genet. 2011, 20, 528–540. [Google Scholar] [CrossRef]
- Rajman, I.; Knapp, L.; Morgan, T.; Masimirembwa, C. African Genetic Diversity: Implications for Cytochrome P450-mediated Drug Metabolism and Drug Development. eBioMedicine 2017, 17, 67–74. [Google Scholar] [CrossRef]
- Dai, D.; Zeldin, D.C.; Blaisdell, J.A.; Chanas, B.; Coulter, S.J.; Ghanayem, B.I.; Goldstein, J.A. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics 2001, 11, 597–607. [Google Scholar] [CrossRef]
- Pratt, V.M.; Scott, S.A.; Pirmohamed, M.; Esquivel, B.; Kattman, B.L.; Malheiro, A.J. Medical Genetics Summaries [Internet]; Bethesda: Washington, DC, USA, 2012. [Google Scholar]
- Kenmogne, V.L.; Takundwa, M.M.; Nweke, E.E.; Monchusi, B.; Dube, P.; Maher, H.; Du Toit, J.; Philip-Cherian, V.; Fru, P.N.; Thimiri Govinda Raj, D.B. The first-in-Africa ex vivo drug sensitivity testing platform identifies novel drug combinations for South African leukaemia patient cohort. Sci. Rep. 2025, 15, 9160. [Google Scholar] [CrossRef]
- Klima, S.; Hurrell, T.; Goolam, M.; Gouws, C.; Engelbrecht, A.M.; Kaur, M.; van den Bout, I. A new dawn: Vitalising translational oncology research in Africa with the help of advanced cell culture models. Transl. Oncol. 2025, 56, 102391. [Google Scholar] [CrossRef]
- Kayalioglu, H.; Patena, J.; Sangeda, R.Z.; Masamu, U.; Mmbando, B.; Njiro, B.; Iyegbe, C.; Gyamfi, J.; Vieira, D.; Peprah, E. The importance of funding and investment to strengthen data science in Africa. Commun. Med. 2025, 5, 293. [Google Scholar] [CrossRef]
- Borges, P.C.C.; Spencer, H.B.; Barbosa, C.; Costa, V.; Furtado, A.; Leal, M.C.; Lopes, C.; Ferreira, D.; Carvalho, A.L.; Dos-Santos-Silva, I.; et al. XPERT® breast cancer STRAT4 as an alternative method of identifying breast cancer phenotype in Cape Verde (preliminary results). Ecancermedicalscience 2023, 17, 1530. [Google Scholar] [CrossRef]
- Alcochete, A. Ethics in Science, Technology and Innovation: Proposed regulation. Braz. J. Clin. Med. Rev. 2023, 1, 19. [Google Scholar] [CrossRef]
- Sariyar, M.; Suhr, S.; Schlunder, I. How Sensitive Is Genetic Data? Biopreserv. Biobank. 2017, 15, 494–501. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, C.S.; Magalhães, A.C.; Pinto, R.J.; Carrilho, C.; Pereira, C.; Miguel, F.; Borges, P.; Santos, L.L.; Pereira, L. Addressing Ancestral Underrepresentation in Oncobiology: The Need for Sub-Saharan African-Specific In Vitro Models. Genes 2025, 16, 1403. https://doi.org/10.3390/genes16121403
dos Santos CS, Magalhães AC, Pinto RJ, Carrilho C, Pereira C, Miguel F, Borges P, Santos LL, Pereira L. Addressing Ancestral Underrepresentation in Oncobiology: The Need for Sub-Saharan African-Specific In Vitro Models. Genes. 2025; 16(12):1403. https://doi.org/10.3390/genes16121403
Chicago/Turabian Styledos Santos, Carla S., Ana C. Magalhães, Ricardo J. Pinto, Carla Carrilho, Cláudia Pereira, Fernando Miguel, Pamela Borges, Lúcio Lara Santos, and Luisa Pereira. 2025. "Addressing Ancestral Underrepresentation in Oncobiology: The Need for Sub-Saharan African-Specific In Vitro Models" Genes 16, no. 12: 1403. https://doi.org/10.3390/genes16121403
APA Styledos Santos, C. S., Magalhães, A. C., Pinto, R. J., Carrilho, C., Pereira, C., Miguel, F., Borges, P., Santos, L. L., & Pereira, L. (2025). Addressing Ancestral Underrepresentation in Oncobiology: The Need for Sub-Saharan African-Specific In Vitro Models. Genes, 16(12), 1403. https://doi.org/10.3390/genes16121403





