Optimizing an Ex Vitro RUBY-Equipped Method for Hairy Root Transformation of Peanuts: An Efficient Approach for the Functional Study of Genes in Peanut Roots
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of Peanuts
2.2. Vector Construction and Transformation
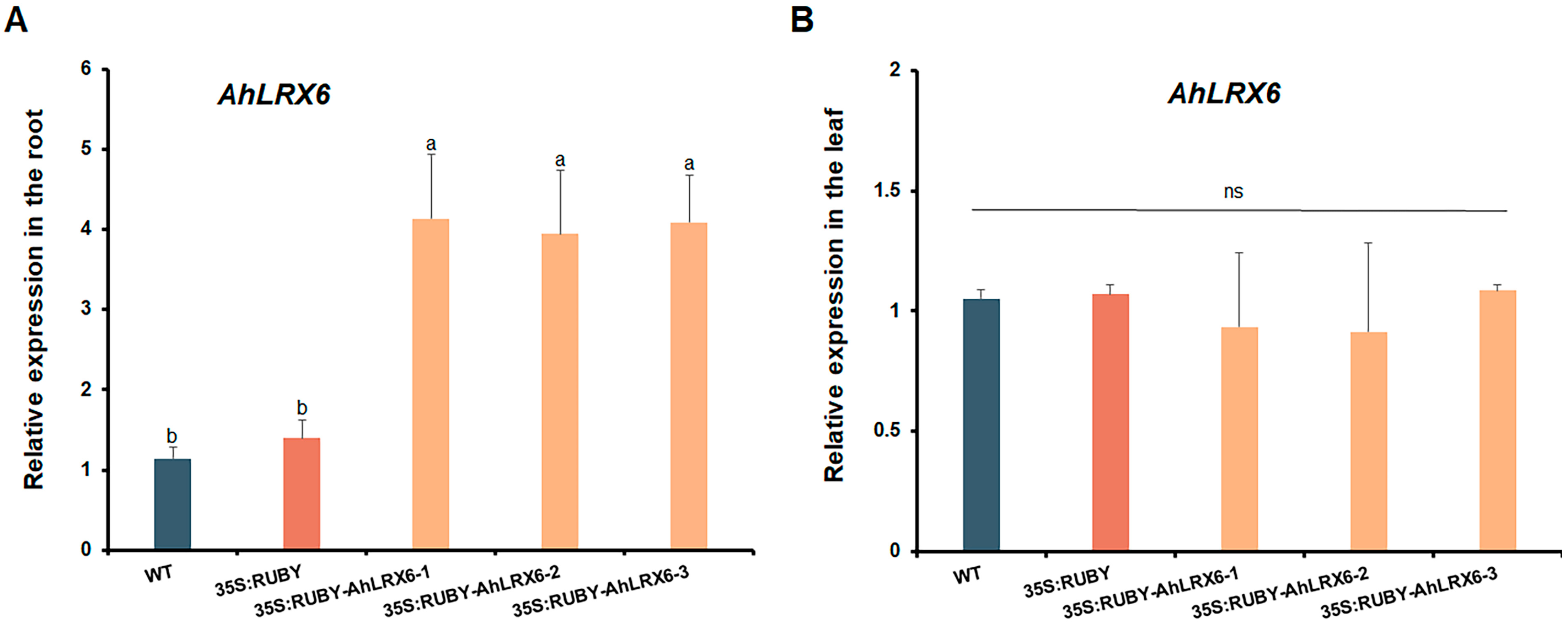
2.3. RT-qPCR Analysis of Peanut Root and Leaf
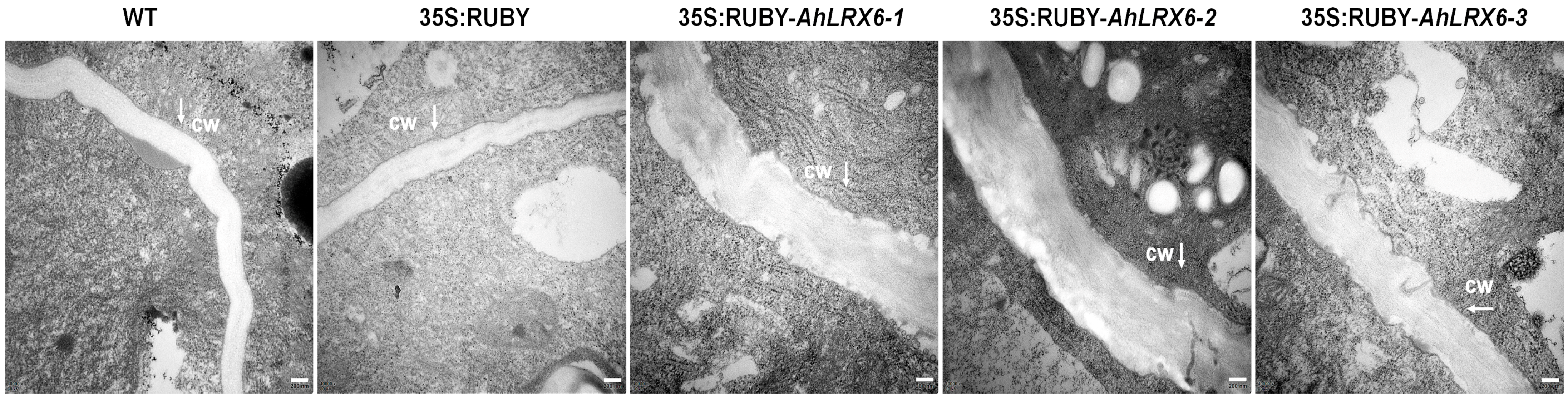
2.4. Transmission Electron Microscopy of Root Tips
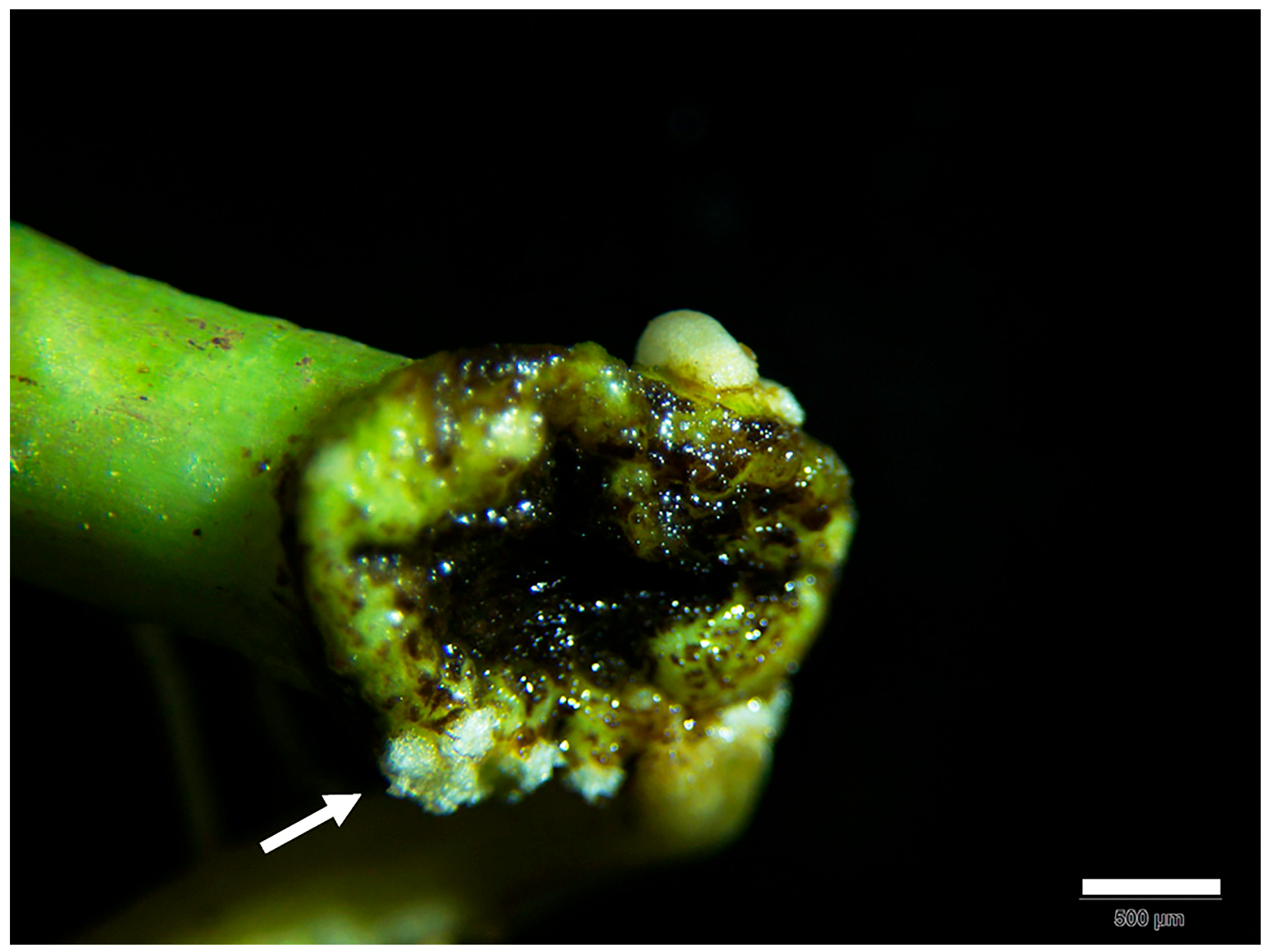
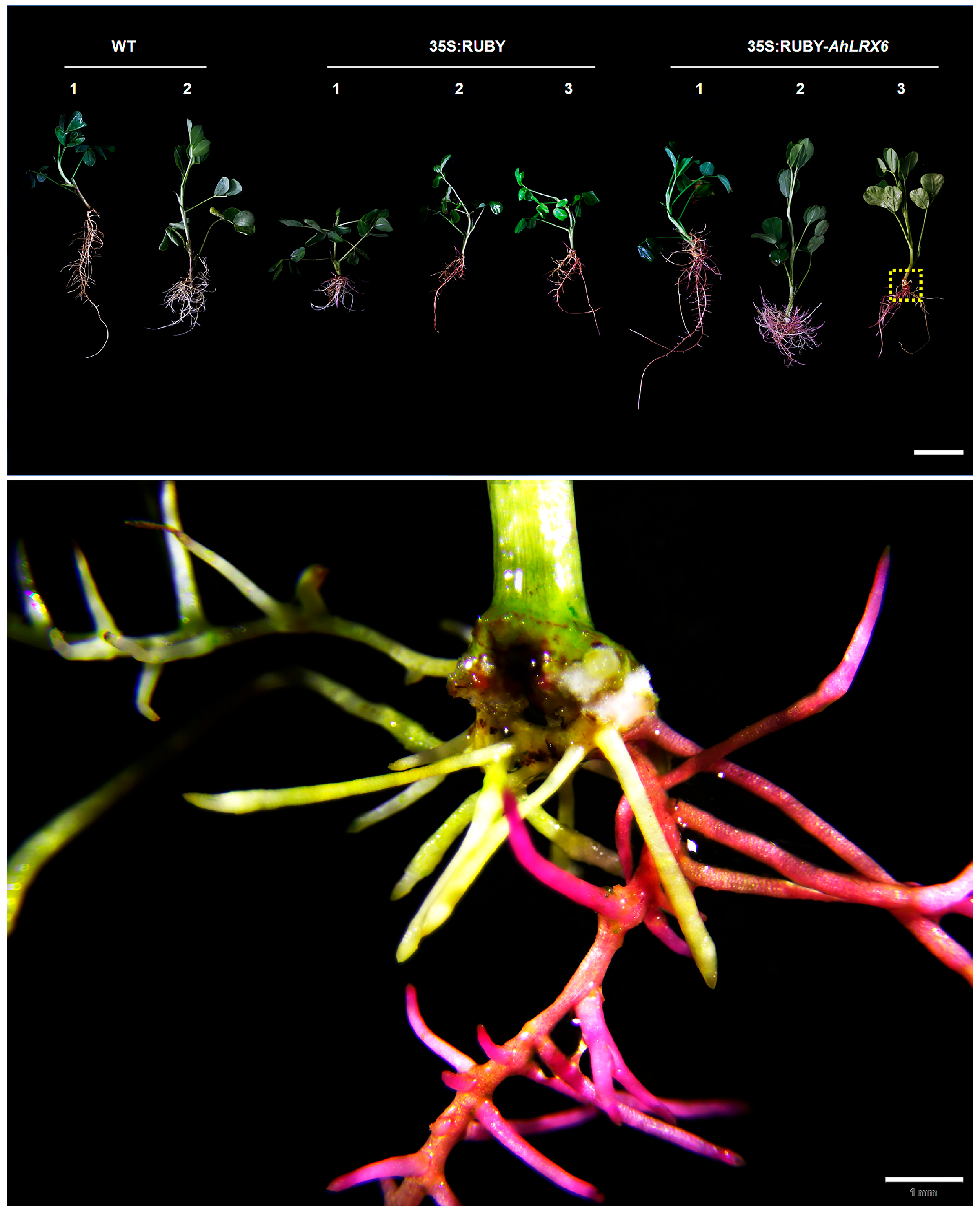
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krishna, G.; Singh, B.K.; Kim, E.K.; Morya, V.K.; Ramteke, P.W. Progress in genetic engineering of peanut (Arachis hypogaea L.)—A review. Plant Biotechnol. J. 2015, 13, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, G.; Rao, T.; Kirti, P.B. Genetic engineering for peanut improvement: Current status and prospects. Plant Cell Tiss. Org. Cul. 2016, 125, 399–416. [Google Scholar] [CrossRef]
- Faraz, R.; Gokhale, M.; Gothalwa, R. Hairy root culture through Agrobacterium rhizogenes for enhancement of secondary metabolites production in medicinal plants: A review. JAMA 2020, 4, 45–58. [Google Scholar] [CrossRef]
- Gutierrez-Valdes, N.; Häkkinen, S.T.; Lemasson, C.; Guillet, M.; Oksman-Caldentey, K.M.; Ritala, A.; Cardon, F. Hairy root cultures-a versatile tool with multiple applications. Front. Plant Sci. 2020, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [PubMed]
- Mecchia, M.A.; Santos-Fernandez, G.; Duss, N.N.; Somoza, S.C.; Boisson-Dernier, A.; Gagliardini, V.; Martínez-Bernardini, A.; Fabrice, T.N.; Ringli, C.; Muschietti, J.P.; et al. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017, 358, 1600–1603. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Deng, S.; Huang, H.; Mo, J.; Xu, Z.F.; Wang, Y. Exploring the Potential Applications of the Noninvasive Reporter Gene RUBY in Plant Genetic Transformation. Forests 2023, 14, 637. [Google Scholar] [CrossRef]
- Ajdanian, L.; Niazian, M.; Torkamaneh, D. Optimizing ex vitro one-step RUBY-equipped hairy root transformation in drug- and hemp-type Cannabis. Plant Biotechnol. J. 2024, 22, 1957–1959. [Google Scholar] [CrossRef] [PubMed]
- Nanjareddy, K.; Zepeda-Jazo, I.; Arthikala, M.K. A protocol for the generation of Arachis hypogaea composite plants: A valuable tool for the functional study of mycorrhizal symbiosis. Appl. Plant Sci. 2022, 10, e11454. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Zhan, J.; Pan, C.; Xiong, W.; Xiao, D.; Wang, Y.; Shen, H.; Wang, A.; He, L. Identification and validation of reference genes for real-time qPCR normalization during Al-induced programmed cell death in peanut. Biol. Plant. 2019, 63, 237–246. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Hong, C.; Lu, W.; Zhang, W.; Gao, H. Quantitative RUBY reporter assay for gene regulation analysis. Plant Cell Environ. 2024, 47, 3701–3711. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Su, L.; Liu, S.; Zeng, X.; Zheng, D.; Hong, L.; Li, L. Agrobacterium rhizogenes-mediated transformation of Arachis hypogaea: An efficient tool for functional study of genes. Biotechnol. Biotechnol. Equip. 2016, 2818, 869–878. [Google Scholar] [CrossRef]
- Guimaraes, L.A.; Pereira, B.M.; Araujo, A.C.G.; Guimaraes, P.M.; Brasileiro, A.C.M. Ex vitro hairy root induction in detached peanut leaves for plant-nematode interaction studies. Plant Methods 2017, 13, 25. [Google Scholar] [PubMed]
- Contag, C.H.; Bachmann, M.H. Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng. 2002, 4, 235–260. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Lu, L.X.; Pan, B.Z.; Chai, X.; Fu, Q.T.; Geng, X.C.; Mo, Y.; Fei, Y.C.; Xu, J.J.; Li, M.; et al. Agrobacterium rhizogenes-Mediated hairy root genetic transformation using Agrobacterium gel inoculation and RUBY reporter enables efficient gene function analysis in Sacha Inchi (Plukenetia volubilis). Int. J. Mol. Sci. 2025, 26, 2496. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Lee, S.Y.; Park, S.U. An efficient protocol for peanut (Arachis hypogaea L.) transformation mediated by Agrobacterium rhizogenes. Rom. Biotechnol. Lett. 2009, 14, 4641–4647. [Google Scholar]
- Akasaka, Y.; Mii, M.; Daimon, H. Morphological alterations and root nodule formation in Agrobacterium rhizogenes-mediated transgenic hairy roots of peanut (Arachis hypogaea L.). Ann. Bot. 1998, 81, 355–362. [Google Scholar] [CrossRef]
- Sinharoy, S.; Saha, S.; Chaudhury, S.R.; Dasgupta, M. Transformed hairy roots of Arachis hypogea: A tool for studying root nodule symbiosis in a non-infection thread legume of the Aeschynomeneae tribe. Mol. Plant Microbe Interact. 2009, 22, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Guimarães, L.A.; Wu, C.L.; Timper, P.; Holbrook, C.C.; Ozias-Akins, P. A technique to study Meloidogyne arenaria resistance in Agrobacterium rhizogenes-transformed peanut. Plant Dis. 2014, 98, 1292–1299. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhou, J.; Kong, F.; Li, X.; Xiao, D.; Wang, A.; He, L.; Zhan, J. Optimizing an Ex Vitro RUBY-Equipped Method for Hairy Root Transformation of Peanuts: An Efficient Approach for the Functional Study of Genes in Peanut Roots. Genes 2025, 16, 1401. https://doi.org/10.3390/genes16121401
Li X, Zhou J, Kong F, Li X, Xiao D, Wang A, He L, Zhan J. Optimizing an Ex Vitro RUBY-Equipped Method for Hairy Root Transformation of Peanuts: An Efficient Approach for the Functional Study of Genes in Peanut Roots. Genes. 2025; 16(12):1401. https://doi.org/10.3390/genes16121401
Chicago/Turabian StyleLi, Xinyue, Jun Zhou, Fei Kong, Xiaoyun Li, Dong Xiao, Aiqin Wang, Longfei He, and Jie Zhan. 2025. "Optimizing an Ex Vitro RUBY-Equipped Method for Hairy Root Transformation of Peanuts: An Efficient Approach for the Functional Study of Genes in Peanut Roots" Genes 16, no. 12: 1401. https://doi.org/10.3390/genes16121401
APA StyleLi, X., Zhou, J., Kong, F., Li, X., Xiao, D., Wang, A., He, L., & Zhan, J. (2025). Optimizing an Ex Vitro RUBY-Equipped Method for Hairy Root Transformation of Peanuts: An Efficient Approach for the Functional Study of Genes in Peanut Roots. Genes, 16(12), 1401. https://doi.org/10.3390/genes16121401




