Dysfunction and Pathological Origins of Lymphatic Endothelial Cells in Atherosclerosis Revealed by Single-Cell Transcriptomics
Abstract
1. Introduction
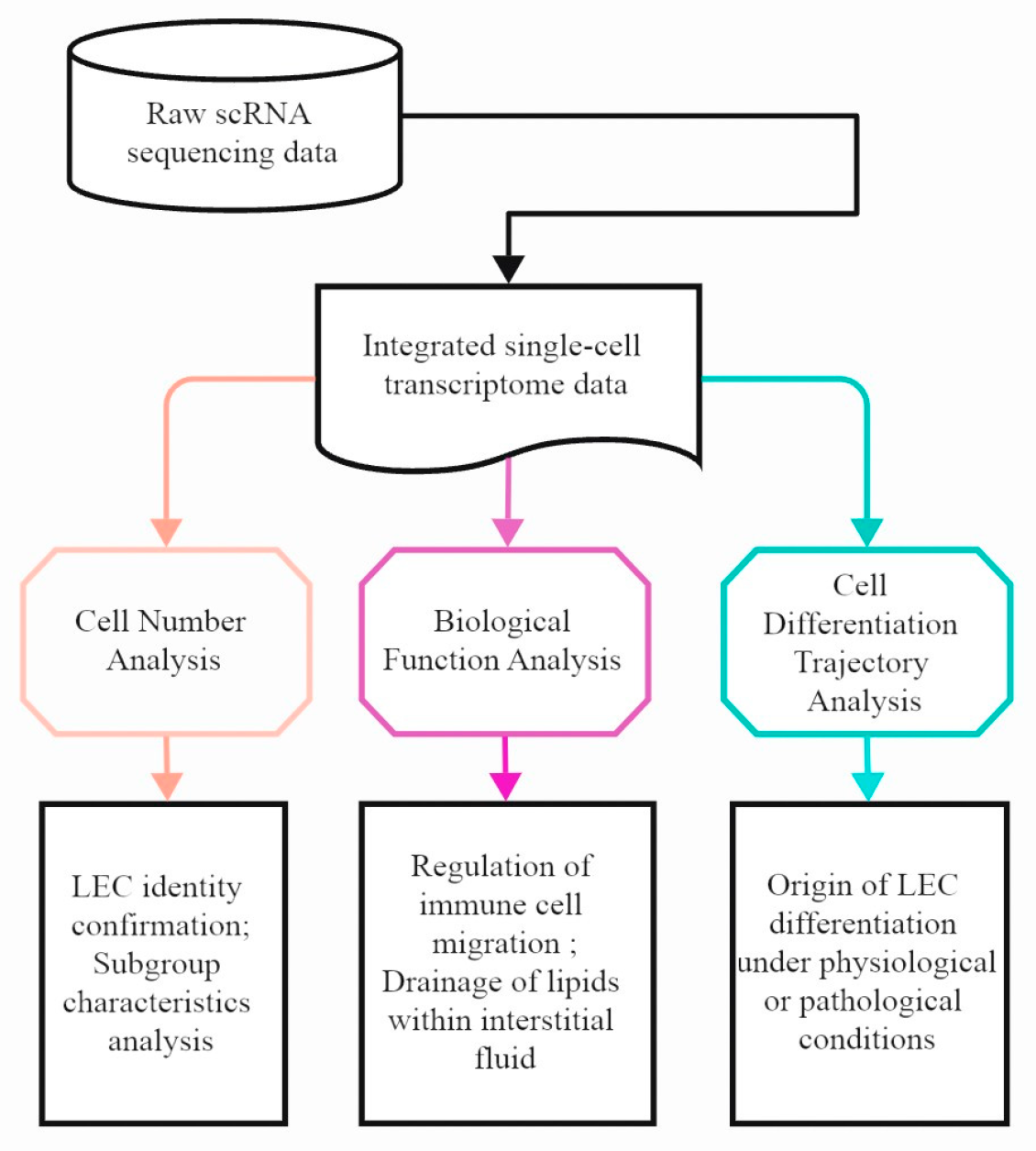
2. Materials and Methods
2.1. Single-Cell RNA-seq Data Acquisition and Processing
2.2. Data Integration Analysis
2.3. Differentially Expressed Genes
2.4. LEC Sub-Clustering
2.5. Gene Signature Score
2.6. Pathway Enrichment Analysis
2.7. Cell-Cell Communication Analysis
2.8. Cell Trajectory Analysis
3. Results
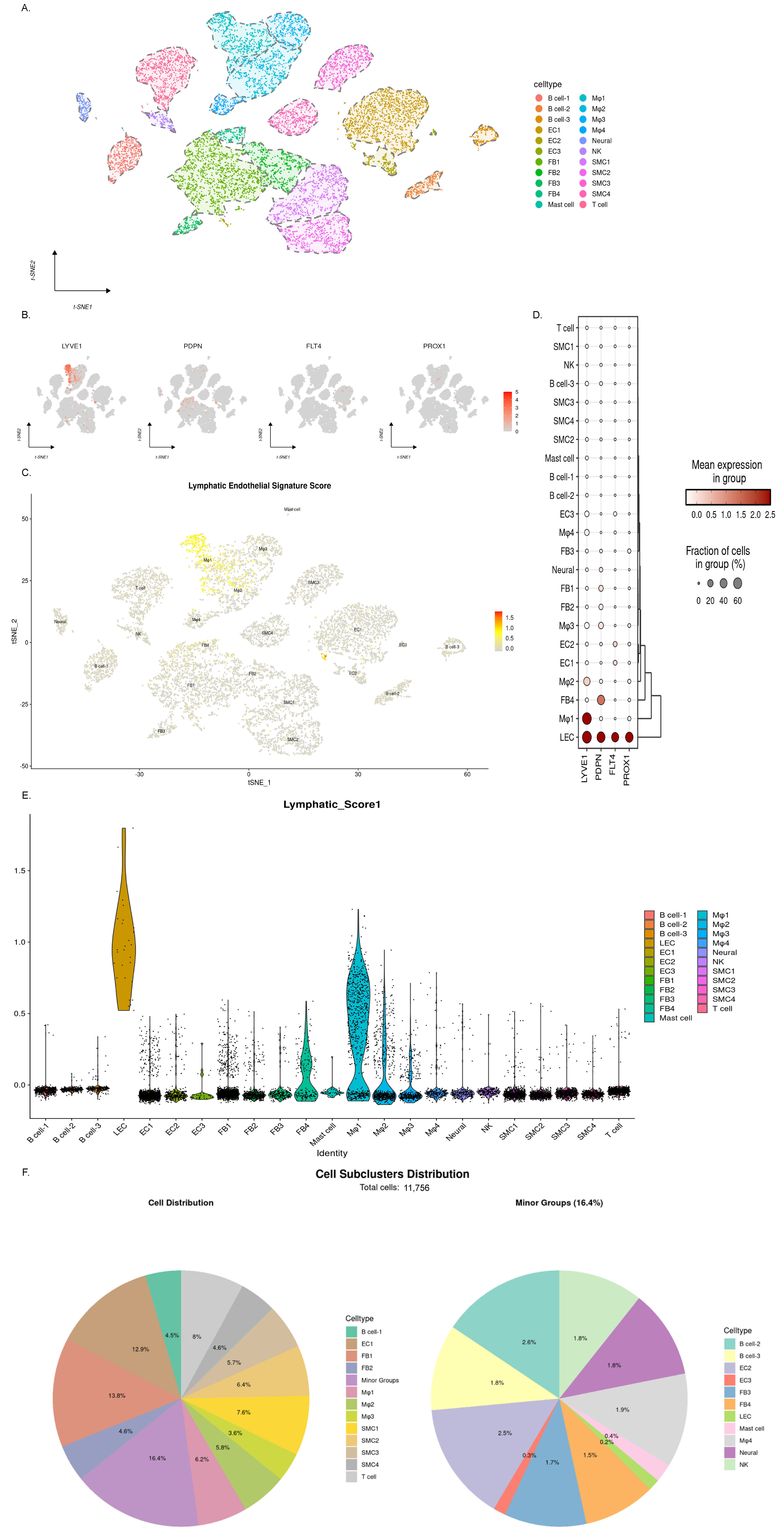
3.1. The Single-Cell RNA Sequencing Results Revealed Endothelial Cell Heterogeneity in Mouse Aortic Atherosclerotic Plaques
3.2. The Expression of Lymphatic-Endothelial-Specific Genes Suggests the Presence of Multiple Cell Populations Associated with Lymphatic Endothelium
3.3. Dynamic Changes in LEC Abundance Suggest Lymphangiogenesis in Early-Stage Atherosclerosis
3.4. Activation of the RAS Signaling Pathway Is a Key Driver of LEC Proliferation
3.5. LEC-Mediated T Cell Migration and Activation in Early Disease Progression
3.6. The Lipo-Transfer Function of LECs Is Disrupted in Atherosclerotic Disease
3.7. The Multi-Differentiated Origin of LECs Under Pathological Conditions Cannot Compensate for the Loss of Function
3.8. Single-Cell Transcriptomic Profiling Reveals a Scarcity of Lymphatic Endothelial Cells in Human Atherosclerotic Coronary Arteries
4. Discussion
Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LEC | Lymphatic endothelial cell |
| VEC | Vascular endothelial cell |
| FB | Fibroblast |
| AS | Atherosclerosis |
References
- Lim, H.Y.; Thiam, C.H.; Yeo, K.P.; Bisoendial, R.; Hii, C.S.; McGrath, K.C.; Tan, K.W.; Heather, A.; Alexander, J.S.J.; Angeli, V. Lymphatic vessels are essential for the removal of cholesterol from peripheral tissues by srbi-mediated transport of hdl. Cell Metab. 2013, 17, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Marston, N.A.; Giugliano, R.P.; Melloni, G.E.M.; Park, J.-G.; Morrill, V.; Blazing, M.A.; Ference, B.; Stein, E.; Stroes, E.S.; Braunwald, E.; et al. Association of Apolipoprotein B–Containing Lipoproteins and Risk of Myocardial Infarction in Individuals with and Without Atherosclerosis: Distinguishing Between Particle Concentration, Type, and Content. JAMA Cardiol. 2022, 7, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Ilyas, I.; Little, P.J.; Li, H.; Kamato, D.; Zheng, X.; Luo, S.; Li, Z.; Liu, P.; Han, J.; et al. Endothelial Dysfunction in Atherosclerotic Cardiovascular Diseases and Beyond: From Mechanism to Pharmacotherapies. Pharmacol. Rev. 2021, 73, 924–967. [Google Scholar] [CrossRef]
- Xue, S.; Su, Z.; Liu, D. Immunometabolism and immune response regulate macrophage function in atherosclerosis. Ageing Res. Rev. 2023, 90, 101993. [Google Scholar] [CrossRef]
- Martel, C.; Li, W.; Fulp, B.; Platt, A.M.; Gautier, E.L.; Westerterp, M.; Bittman, R.; Tall, A.R.; Chen, S.-H.; Thomas, M.J.; et al. Lymphatic vasculature mediates macrophage reverse cholesterol transport in mice. J. Clin. Investig. 2013, 123, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Yeo, K.P.; Lim, H.Y.; Thiam, C.H.; Azhar, S.H.; Tan, C.; Tang, Y.; See, W.Q.; Koh, X.H.; Zhao, M.H.; Phua, M.L.; et al. Efficient aortic lymphatic drainage is necessary for atherosclerosis regression induced by ezetimibe. Sci. Adv. 2020, 6, eabc2697. [Google Scholar] [CrossRef]
- Milasan, A.; Smaani, A.; Martel, C. Early rescue of lymphatic function limits atherosclerosis progression in Ldlr-/- mice. Atherosclerosis 2019, 283, 106–119. [Google Scholar] [CrossRef]
- Brakenhielm, E.; Alitalo, K. Cardiac lymphatics in health and disease. Nat. Rev. Cardiol. 2019, 16, 56–68. [Google Scholar] [CrossRef]
- Wirka, R.C.; Wagh, D.; Paik, D.T.; Pjanic, M.; Nguyen, T.; Miller, C.L.; Kundu, R.; Nagao, M.; Coller, J.; Koyano, T.K.; et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 2019, 25, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Plikus, M.V.; Nie, Q. CellChat for systematic analysis of cell-cell communication from single-cell transcriptomics. Nat. Protoc. 2025, 20, 180–219. [Google Scholar] [CrossRef]
- Lu, H.; Ping, J.; Zhou, G.; Zhao, Z.; Gao, W.; Jiang, Y.; Quan, C.; Lu, Y.; Zhou, G. CommPath: An R package for inference and analysis of pathway-mediated cell-cell communication chain from single-cell transcriptomics. Comput. Struct. Biotechnol. J. 2022, 20, 5978–5983. [Google Scholar] [CrossRef] [PubMed]
- Street, K.; Risso, D.; Fletcher, R.B.; Das, D.; Ngai, J.; Yosef, N.; Purdom, E.; Dudoit, S. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018, 19, 477. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, N.; He, Y.; D’Addio, M.; Tacconi, C.; Detmar, M.; Dieterich, L.C. Single-cell mapping reveals new markers and functions of lymphatic endothelial cells in lymph nodes. PLoS Biol. 2020, 18, e3000704. [Google Scholar] [CrossRef]
- Taher, M.; Nakao, S.; Zandi, S.; Melhorn, M.I.; Hayes, K.C.; Hafezi-Moghadam, A. Phenotypic transformation of intimal and adventitial lymphatics in atherosclerosis: A regulatory role for soluble VEGF receptor 2. FASEB J. 2016, 30, 2490–2499. [Google Scholar] [CrossRef]
- Feng, X.; Du, M.; Li, S.; Zhang, Y.; Ding, J.; Wang, J.; Wang, Y.; Liu, P. Hydroxysafflor yellow A regulates lymphangiogenesis and inflammation via the inhibition of PI3K on regulating AKT/mTOR and NF-κB pathway in macrophages to reduce atherosclerosis in ApoE-/- mice. Phytomedicine 2023, 112, 154684. [Google Scholar] [CrossRef]
- Petrova, T.V.; Koh, G.Y. Biological functions of lymphatic vessels. Science 2020, 369, eaax4063. [Google Scholar] [CrossRef]
- Liu, X.; Cui, K.; Wu, H.; Li, K.S.; Peng, Q.; Wang, D.; Cowan, D.B.; Dixon, J.B.; Srinivasan, R.S.; Bielenberg, D.R.; et al. Promoting Lymphangiogenesis and Lymphatic Growth and Remodeling to Treat Cardiovascular and Metabolic Diseases. Arterioscler. Thromb. Vasc. Biol. 2023, 43, e1–e10. [Google Scholar] [CrossRef]
- Miller, C.N.; Hartigan-O’cOnnor, D.J.; Lee, M.S.; Laidlaw, G.; Cornelissen, I.P.; Matloubian, M.; Coughlin, S.R.; McDonald, D.M.; McCune, J.M. IL-7 production in murine lymphatic endothelial cells and induction in the setting of peripheral lymphopenia. Int. Immunol. 2013, 25, 471–483. [Google Scholar] [CrossRef]
- Lyu, Q.; Ley, K. How Lymphatic Endothelial Cells Destabilize Regulatory T Cells. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 215–217. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.; Lu, S.; Zhang, C.; Ma, Z.; Su, R.; Li, Y.; Sun, T.; Li, Y.; Hong, M.; et al. Pairing of single-cell RNA analysis and T cell antigen receptor profiling indicates breakdown of T cell tolerance checkpoints in atherosclerosis. Nat. Cardiovasc. Res. 2023, 2, 290–306. [Google Scholar] [CrossRef]
- Ulvmar, M.H.; Mäkinen, T. Heterogeneity in the lymphatic vascular system and its origin. Cardiovasc. Res. 2016, 111, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Escobedo, N.; Oliver, G. Lymphangiogenesis: Origin, Specification, and Cell Fate Determination. Annu. Rev. Cell Dev. Biol. 2016, 32, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Klotz, L.; Norman, S.; Vieira, J.M.; Masters, M.; Rohling, M.; Dubé, K.N.; Bollini, S.; Matsuzaki, F.; Carr, C.A.; Riley, P.R. Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature 2015, 522, 62–67. [Google Scholar] [CrossRef]
- Marziano, C.; Genet, G.; Hirschi, K.K. Vascular endothelial cell specification in health and disease. Angiogenesis 2021, 24, 213–236. [Google Scholar] [CrossRef]
- Ni, Q.; Li, G.; Chen, Y.; Bao, C.; Wang, T.; Li, Y.; Ruan, X.; Wang, H.; Sun, W. LECs regulate neutrophil clearance through IL-17RC/CMTM4/NF-κB axis at sites of inflammation or infection. Mucosal Immunol. 2024, 17, 723–738. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Q.; Gu, G.; Yang, D.; Zheng, Y. Dysfunction and Pathological Origins of Lymphatic Endothelial Cells in Atherosclerosis Revealed by Single-Cell Transcriptomics. Genes 2025, 16, 1398. https://doi.org/10.3390/genes16121398
Shen Q, Gu G, Yang D, Zheng Y. Dysfunction and Pathological Origins of Lymphatic Endothelial Cells in Atherosclerosis Revealed by Single-Cell Transcriptomics. Genes. 2025; 16(12):1398. https://doi.org/10.3390/genes16121398
Chicago/Turabian StyleShen, Qinhang, Guangchao Gu, Dan Yang, and Yuehong Zheng. 2025. "Dysfunction and Pathological Origins of Lymphatic Endothelial Cells in Atherosclerosis Revealed by Single-Cell Transcriptomics" Genes 16, no. 12: 1398. https://doi.org/10.3390/genes16121398
APA StyleShen, Q., Gu, G., Yang, D., & Zheng, Y. (2025). Dysfunction and Pathological Origins of Lymphatic Endothelial Cells in Atherosclerosis Revealed by Single-Cell Transcriptomics. Genes, 16(12), 1398. https://doi.org/10.3390/genes16121398






