Integrated Bioinformatics and Machine Learning for Ascertainment and Validation of Biomarkers for Screening Breast Disease
Abstract
1. Introduction
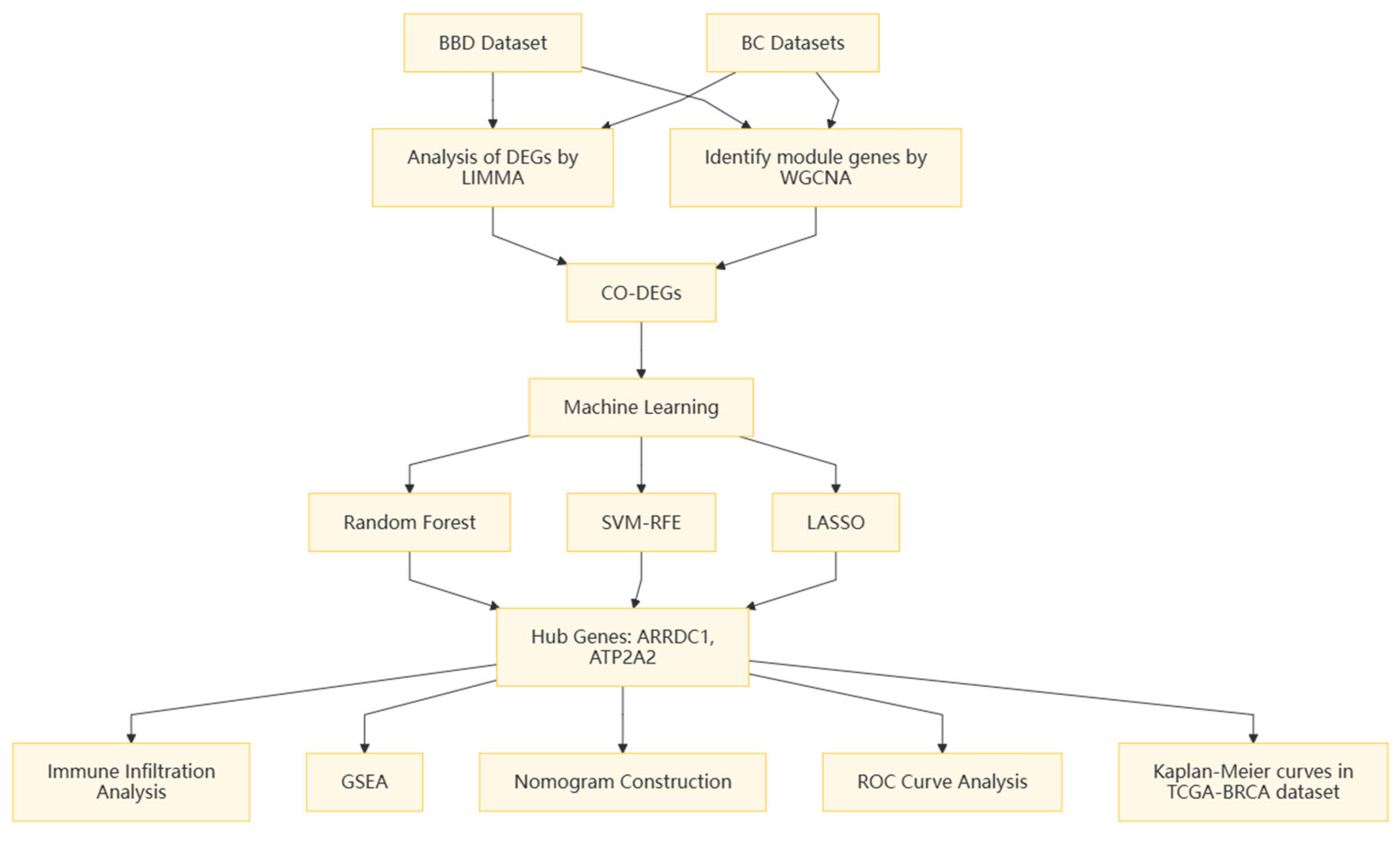
2. Methods
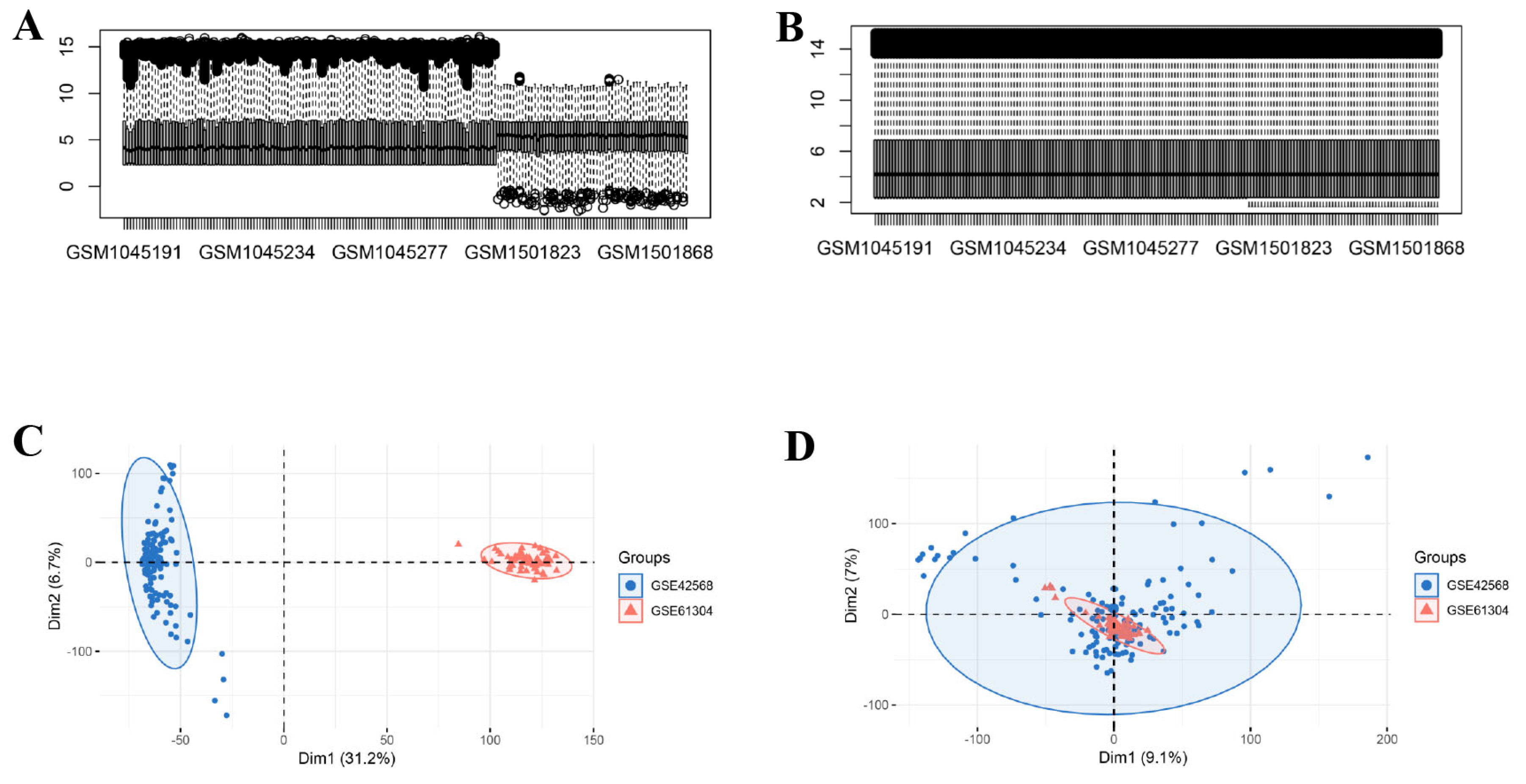
2.1. Microarray Data
2.2. Differential Expression and Enrichment Analysis
2.3. WGCNA
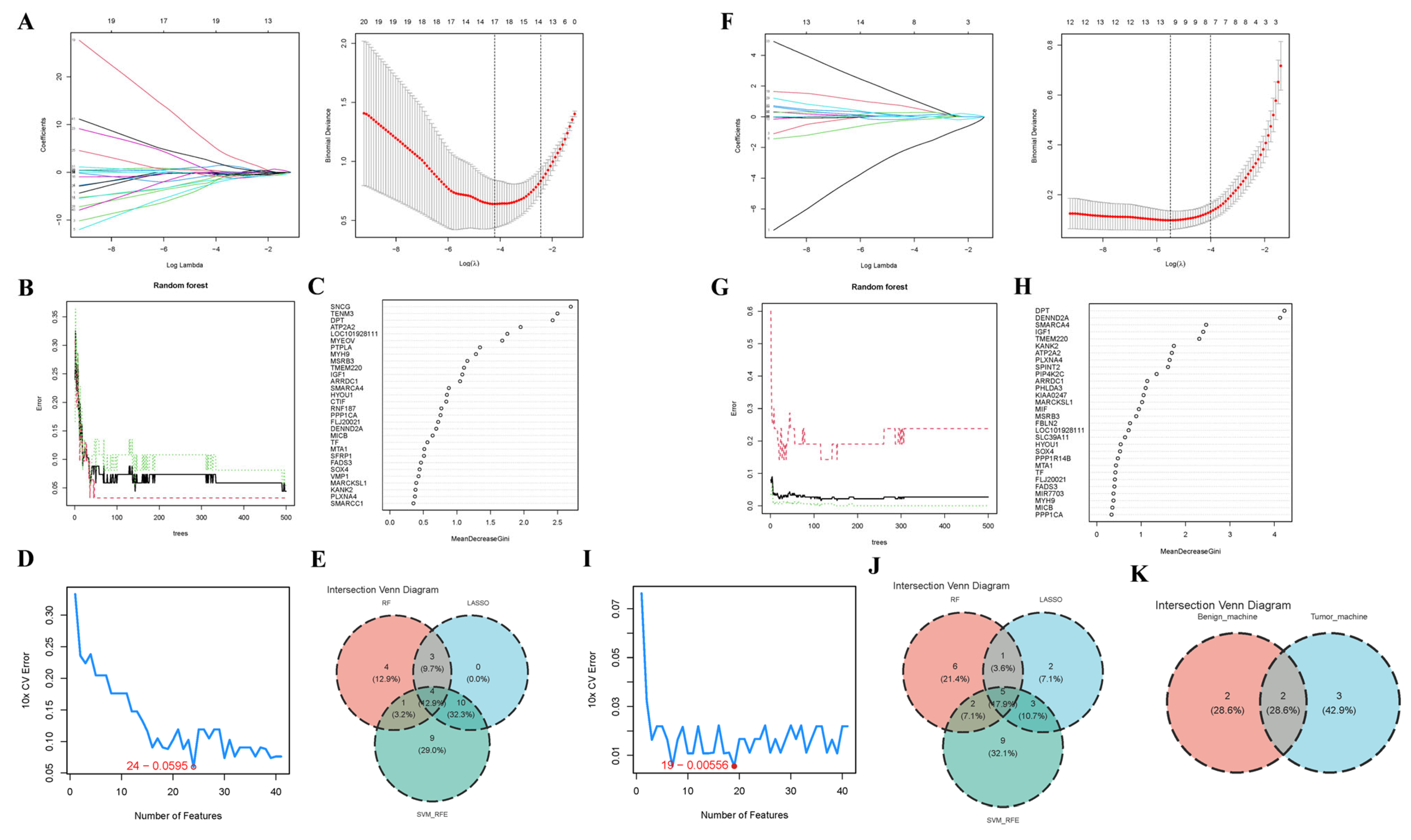
2.4. Core Gene Screening
2.5. Immune Infiltration Analysis
2.6. Statistical Analysis
3. Results
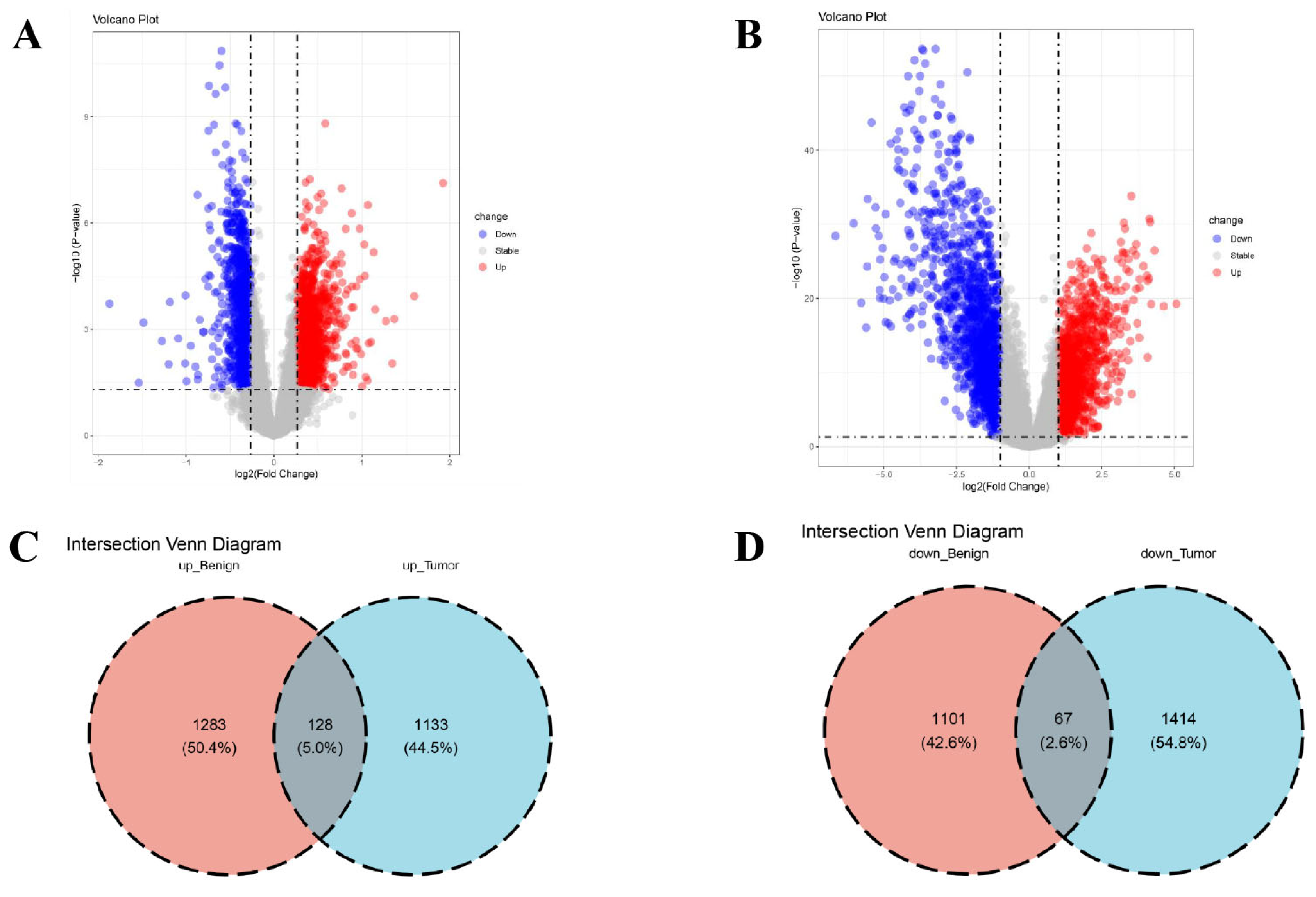
3.1. Identification of DEGs
3.2. Enrichment Analysis of DEGs
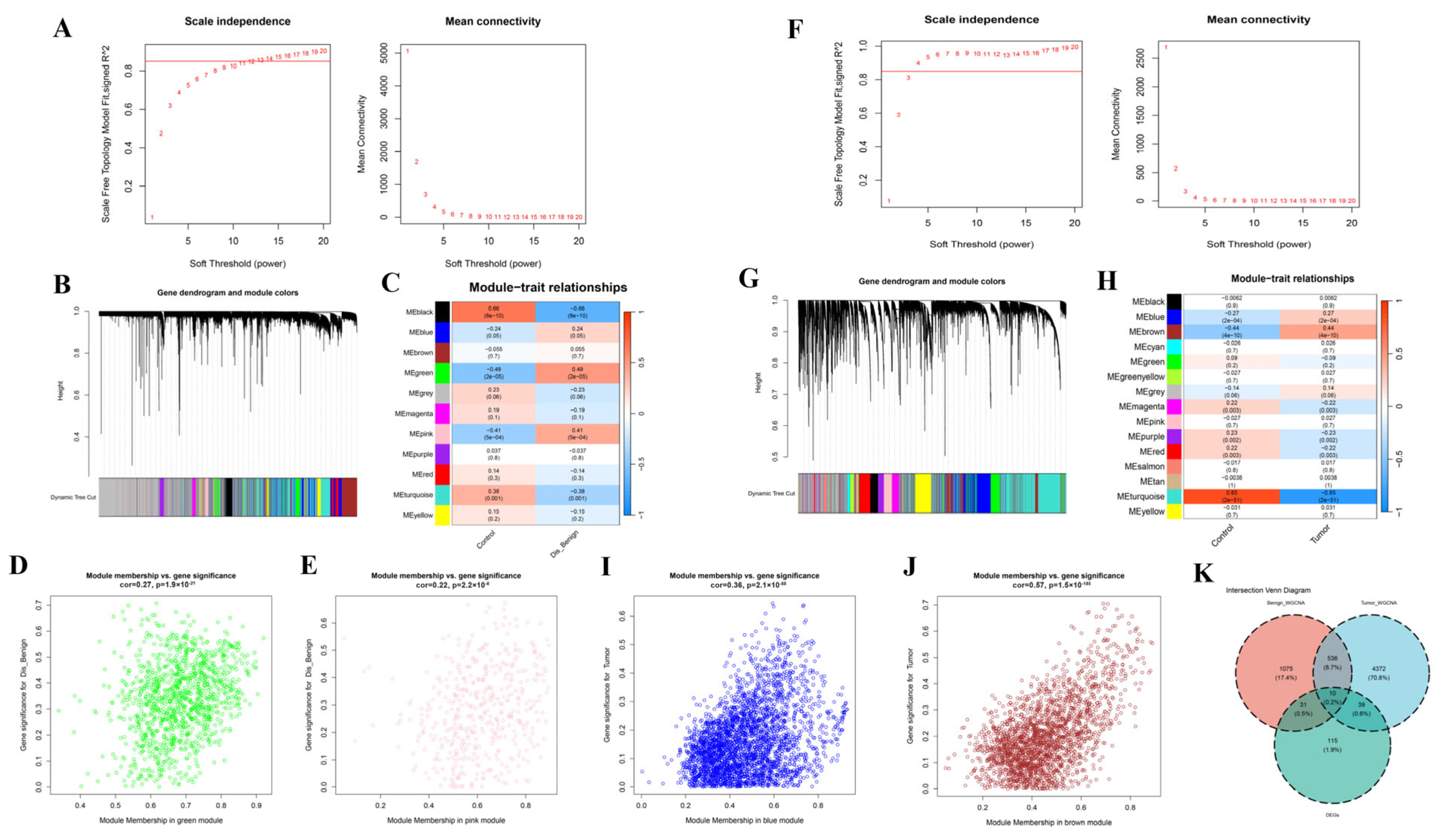
3.3. WGCNA Screening of Key Modules
3.4. Core Gene Screening Based upon ML
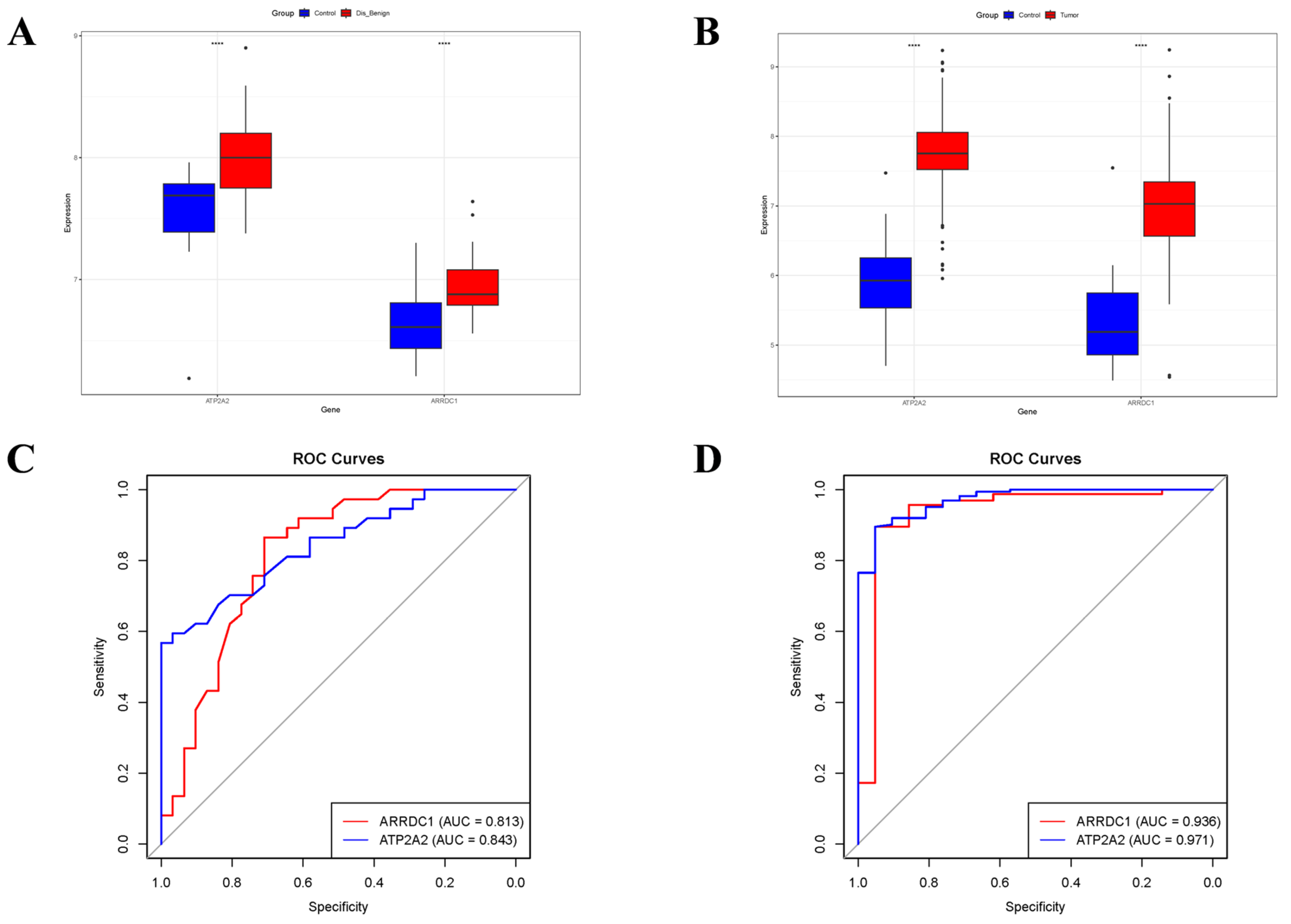
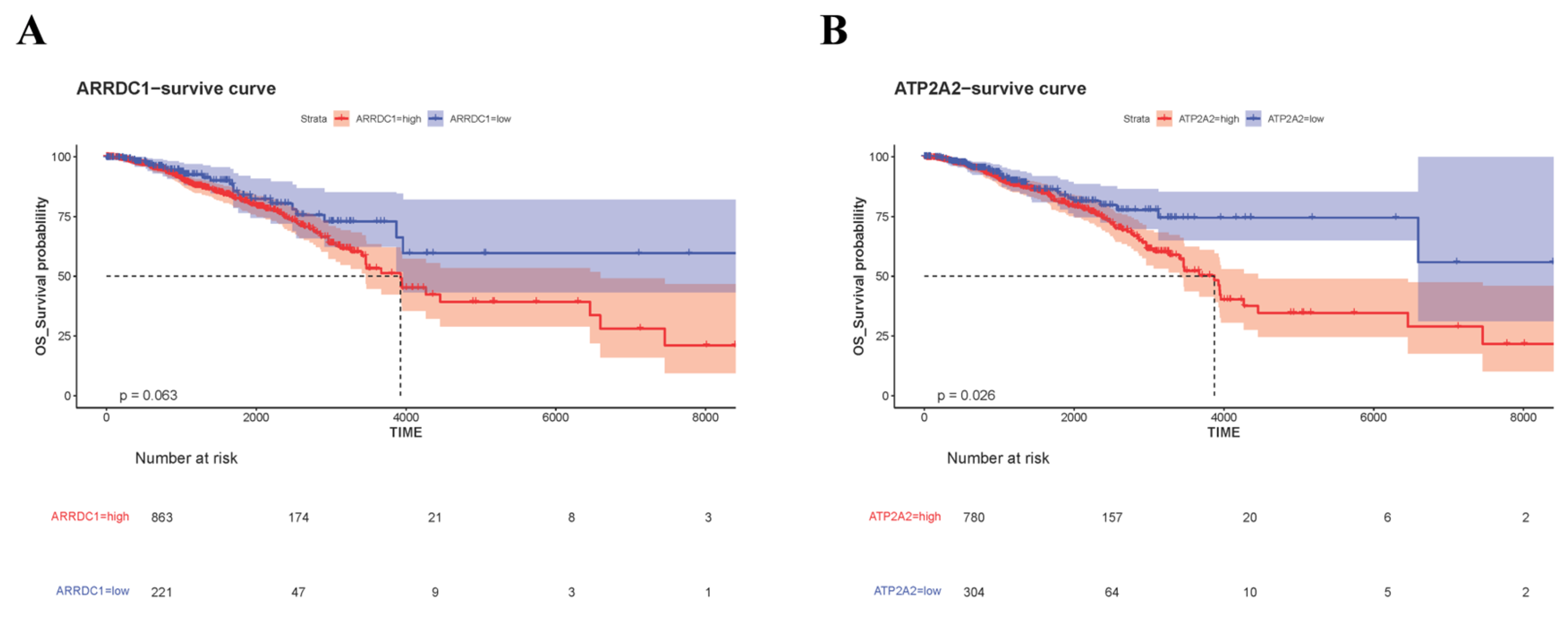
3.5. Validation of Core Genes
3.6. Construction of the Nomogram
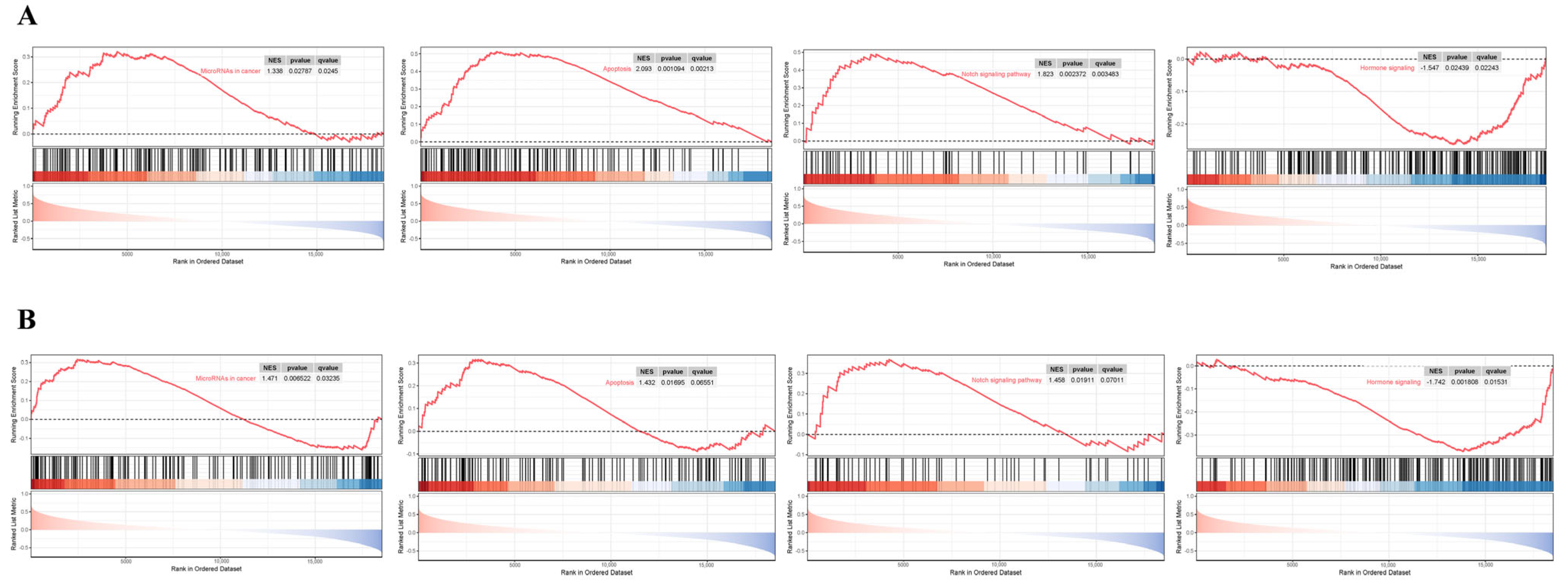
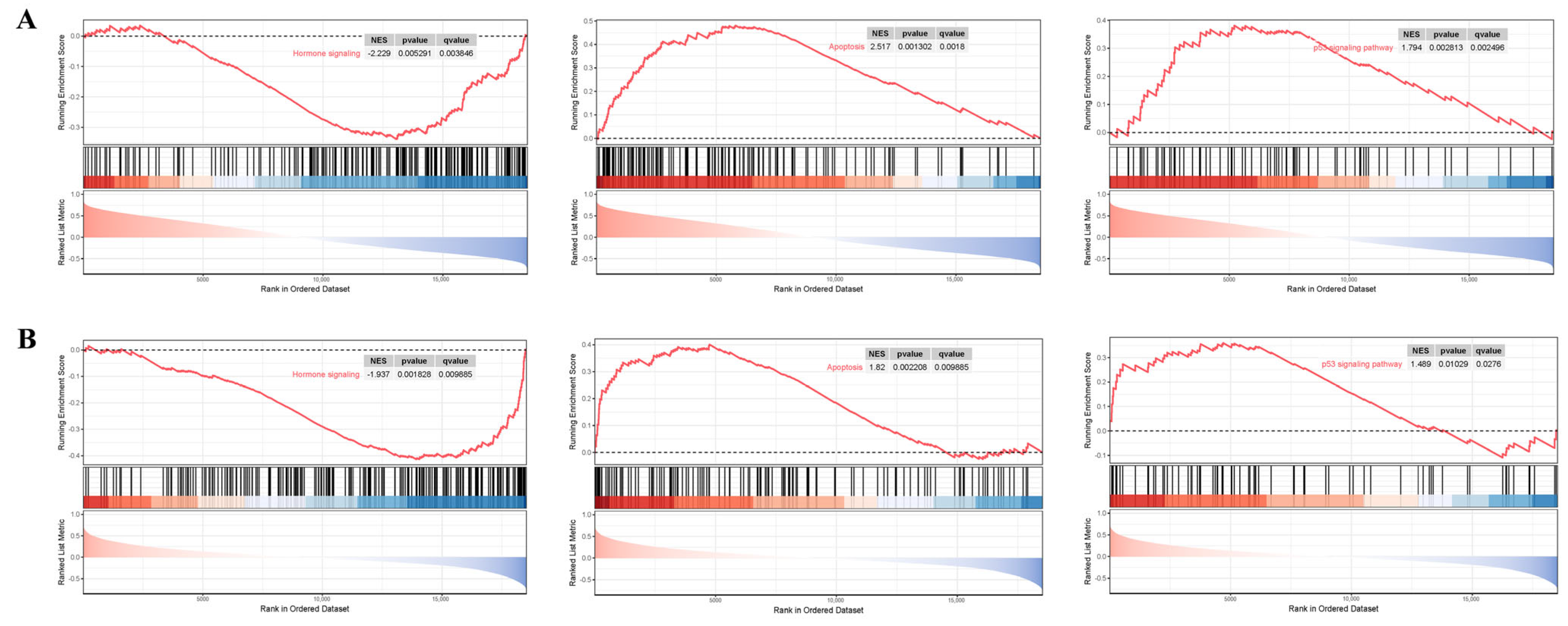
3.7. GSEA Analysis
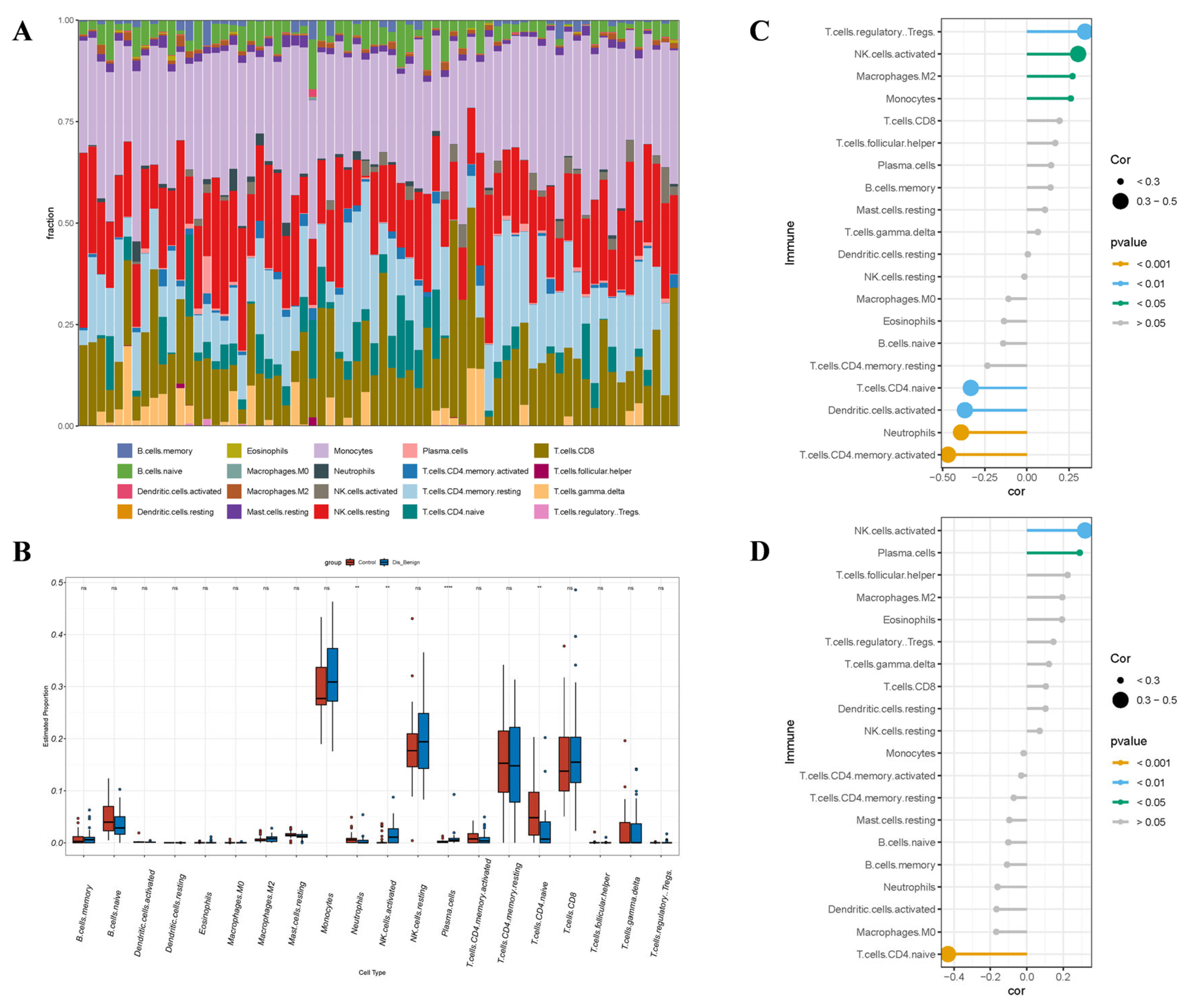
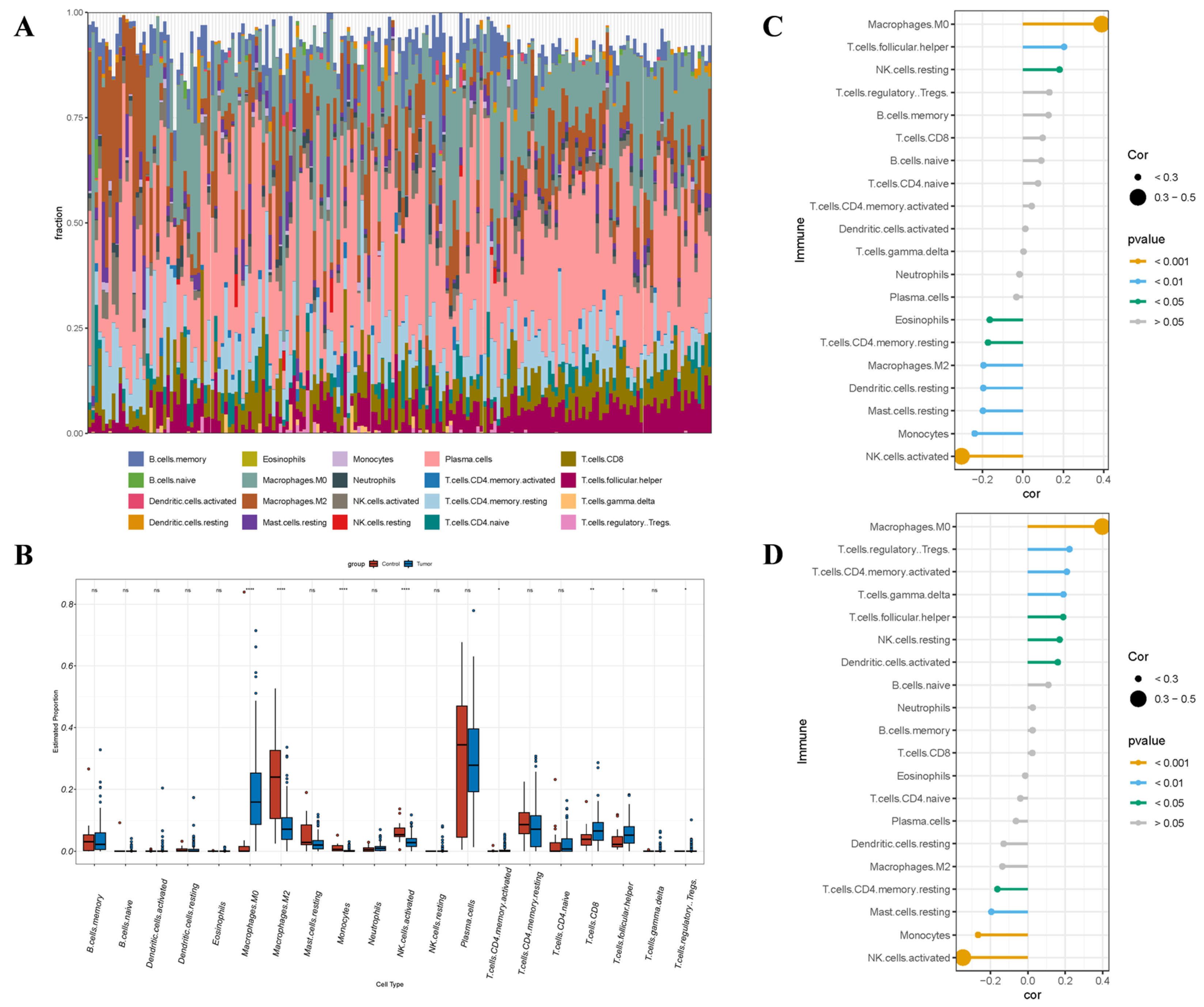
3.8. Immunoanalysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J Clin 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Lee, A.; Lam, C.W. Application of Metabolic Biomarkers in Breast Cancer: A Literature Review. Ann. Lab. Med. 2025, 45, 229–246. [Google Scholar] [CrossRef]
- Weaver, M.; Stuckey, A. Benign Breast Disorders. Obstet. Gynecol. Clin. N. Am. 2022, 49, 57–72. [Google Scholar] [CrossRef]
- Carter, C.L.; Corle, D.K.; Micozzi, M.S.; Schatzkin, A.; Taylor, P.R. A prospective study of the development of breast cancer in 16,692 women with benign breast disease. Am. J. Epidemiol. 1988, 128, 467–477. [Google Scholar] [CrossRef]
- Dupont, W.D.; Parl, F.F.; Hartmann, W.H.; Brinton, L.A.; Winfield, A.C.; Worrell, J.A.; Schuyler, P.A.; Plummer, W.D. Breast cancer risk associated with proliferative breast disease and atypical hyperplasia. Cancer 1993, 71, 1258–1265. [Google Scholar] [CrossRef]
- Johansson, A.; Christakou, A.E.; Iftimi, A.; Eriksson, M.; Tapia, J.; Skoog, L.; Benz, C.C.; Rodriguez-Wallberg, K.A.; Hall, P.; Czene, K.; et al. Characterization of Benign Breast Diseases and Association With Age, Hormonal Factors, and Family History of Breast Cancer Among Women in Sweden. JAMA Netw. Open 2021, 4, e2114716. [Google Scholar] [CrossRef]
- Ma, W.; Jin, Q.; Wu, Y.; Jin, F. Expert Consensus on Diagnosis and Treatment of Breast Hyperplasia. Chin. J. Pract. Surg. 2016, 36, 759–762. [Google Scholar]
- Lilleborge, M.; Falk, R.S.; Sørlie, T.; Ursin, G.; Hofvind, S. Can breast cancer be stopped? Modifiable risk factors of breast cancer among women with a prior benign or premalignant lesion. Int. J. Cancer 2021, 149, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.; Madden, S.F.; Doolan, P.; Aherne, S.T.; Joyce, H.; O’Driscoll, L.; Gallagher, W.M.; Hennessy, B.T.; Moriarty, M.; Crown, J.; et al. Correlating transcriptional networks to breast cancer survival: A large-scale coexpression analysis. Carcinogenesis 2013, 34, 2300–2308. [Google Scholar] [CrossRef] [PubMed]
- LaBreche, H.G.; Nevins, J.R.; Huang, E. Integrating factor analysis and a transgenic mouse model to reveal a peripheral blood predictor of breast tumors. BMC Med. Genom. 2011, 4, 61. [Google Scholar] [CrossRef]
- Grinchuk, O.V.; Motakis, E.; Yenamandra, S.P.; Ow, G.S.; Jenjaroenpun, P.; Tang, Z.; Yarmishyn, A.A.; Ivshina, A.V.; Kuznetsov, V.A. Sense-antisense gene-pairs in breast cancer and associated pathological pathways. Oncotarget 2015, 6, 42197–42221. [Google Scholar] [CrossRef]
- Aswad, L.; Yenamandra, S.P.; Ow, G.S.; Grinchuk, O.; Ivshina, A.V.; Kuznetsov, V.A. Genome and transcriptome delineation of two major oncogenic pathways governing invasive ductal breast cancer development. Oncotarget 2015, 6, 36652–36674. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jensen, M.A.; Zenklusen, J.C. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol. Biol. 2016, 1418, 111–141. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.G.O. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Wu, S.Y.; Fu, T.; Jiang, Y.Z.; Shao, Z.M. Natural killer cells in cancer biology and therapy. Mol. Cancer 2020, 19, 120. [Google Scholar] [CrossRef]
- Vivier, E.; Raulet, D.H.; Moretta, A.; Caligiuri, M.A.; Zitvogel, L.; Lanier, L.L.; Yokoyama, W.M.; Ugolini, S. Innate or adaptive immunity? The example of natural killer cells. Science 2011, 331, 44–49. [Google Scholar] [CrossRef]
- Jokhadze, N.; Das, A.; Dizon, D.S. Global cancer statistics: A healthy population relies on population health. CA Cancer J. Clin. 2024, 74, 224–226. [Google Scholar] [CrossRef]
- Santaguida, S.; Richardson, A.; Iyer, D.R.; M’Saad, O.; Zasadil, L.; Knouse, K.A.; Wong, Y.L.; Rhind, N.; Desai, A.; Amon, A. Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Dev. Cell 2017, 41, 638–651.e635. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Bhat, G.R.; Sethi, I.; Sadida, H.Q.; Rah, B.; Mir, R.; Algehainy, N.; Albalawi, I.A.; Masoodi, T.; Subbaraj, G.K.; Jamal, F.; et al. Cancer cell plasticity: From cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance. Cancer Metastasis Rev. 2024, 43, 197–228. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, E.A.; Sutherland, R.L. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 2009, 9, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Yang, X.; Chen, C.; He, J.; Liang, Y.; Luo, X.; Huang, C.; Wu, W.; Zhang, P.; Liu, C. ARRDC1 inhibits the replication of Semliki Forest virus by regulating the ubiquitination and degradation of viral nsP4. J. Virol. 2025, 99, e0097725. [Google Scholar] [CrossRef]
- Rauch, S.; Martin-Serrano, J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: Implications for PPXY-dependent budding. J. Virol. 2011, 85, 3546–3556. [Google Scholar] [CrossRef]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef]
- Zbieralski, K.; Wawrzycka, D. α-Arrestins and Their Functions: From Yeast to Human Health. Int. J. Mol. Sci. 2022, 23, 4988. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, Y.; Zhou, G.; Qin, G.; Tan, C.; Yin, T.; Zhao, D.; Yao, S. Age-stratified proteomic characteristics and identification of promising precise clinical treatment targets of colorectal cancer. J. Proteom. 2023, 277, 104863. [Google Scholar] [CrossRef]
- Hou, P.P.; Luo, L.J.; Chen, H.Z.; Chen, Q.T.; Bian, X.L.; Wu, S.F.; Zhou, J.X.; Zhao, W.X.; Liu, J.M.; Wang, X.M.; et al. Ectosomal PKM2 Promotes HCC by Inducing Macrophage Differentiation and Remodeling the Tumor Microenvironment. Mol. Cell 2020, 78, 1192–1206.e1110. [Google Scholar] [CrossRef]
- Li, M.; Lin, C.; Cai, Z. Breast cancer stem cell-derived extracellular vesicles transfer ARRDC1-AS1 to promote breast carcinogenesis via a miR-4731-5p/AKT1 axis-dependent mechanism. Transl. Oncol. 2023, 31, 101639. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Koirala, P.; Ding, X.; Chen, B.; Wang, Y.; Wang, Z.; Wang, C.; Zhang, X.; Mo, Y.Y. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget 2016, 7, 20584–20596. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhong, N.; Li, W.; Liu, H.; Yu, H.; Li, Y. Research Progress on the Role of ATP2A2 in Tumor Occurrence Mechanism via Calcium Ion Concentration Changes. J. Second. Mil. Med. Univ. 2013, 34, 1248–1252. [Google Scholar] [CrossRef]
- Korosec, B.; Glavac, D.; Rott, T.; Ravnik-Glavac, M. Alterations in the ATP2A2 gene in correlation with colon and lung cancer. Cancer Genet. Cytogenet. 2006, 171, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Yang, Y.M.; Kim, H.S.; Lee, S.I.; Muallem, S.; Shin, D.M. Markers of squamous cell carcinoma in sarco/endoplasmic reticulum Ca2+ ATPase 2 heterozygote mice keratinocytes. Prog. Biophys. Mol. Biol. 2010, 103, 81–87. [Google Scholar] [CrossRef]
- Atzmony, L.; Zagairy, F.; Mawassi, B.; Shehade, M.; Tatour, Y.; Danial-Farran, N.; Khayat, M.; Warrour, N.; Dodiuk-Gad, R.; Cohen-Barak, E. Persistent Cutaneous Lesions of Darier Disease and Second-Hit Somatic Variants in ATP2A2 Gene. JAMA Dermatol. 2024, 160, 518–524. [Google Scholar] [CrossRef]
- Wang, Q.; Karvelsson, S.T.; Johannsson, F.; Vilhjalmsson, A.I.; Hagen, L.; de Miranda Fonseca, D.; Sharma, A.; Slupphaug, G.; Rolfsson, O. UDP-glucose dehydrogenase expression is upregulated following EMT and differentially affects intracellular glycerophosphocholine and acetylaspartate levels in breast mesenchymal cell lines. Mol. Oncol. 2022, 16, 1816–1840. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, Y.; Liu, R.; Hamdy, H.; Shi, Z.; Jiang, D.; Sun, J. The role of tumor microenvironment and signaling pathways in regulating breast cancer stem cells: Implications for therapy resistance and tumor recurrence. Drug Resist. Updat. 2025, 84, 101315. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Y.; Lei, F.; Luo, A.; Jing, X.; Wang, L.; Lei, J.; Shao, S. Correlation between Notch signaling pathway and YAP1/TAZ in breast cancer. J. Xi’an Jiaotong Univ. (Med. Sci.) 2020, 41, 888–895. [Google Scholar]
- Shah Hosseini, R.; Nouri, S.M.; Bansal, P.; Hjazi, A.; Kaur, H.; Hussein Kareem, A.; Kumar, A.; Al Zuhairi, R.A.H.; Al-Shaheri, N.A.; Mahdavi, P. The p53/miRNA Axis in Breast Cancer. DNA Cell Biol. 2024, 43, 549–558. [Google Scholar] [CrossRef]
- Michmerhuizen, A.R.; Spratt, D.E.; Pierce, L.J.; Speers, C.W. ARe we there yet? Understanding androgen receptor signaling in breast cancer. NPJ Breast Cancer 2020, 6, 47. [Google Scholar] [CrossRef]


| Dataset | Sample Size | Platform | Note | |
|---|---|---|---|---|
| GSE27562 | Healthy control 31 | Benign breast abnormalities 37 | GPL570 | Test dataset |
| GSE42568 | Healthy control 17 | Breast cancer 104 | GPL570 | Test dataset |
| GSE61304 | Healthy control 4 | Breast cancer 58 | GPL570 | Test dataset |
| TCGA-BRCA | Paracancerous 113 | Breast cancer 1106 | TCGA | Validation dataset |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Yang, S.; Zhang, Y.; Piao, C.; Liu, X.; Wu, X. Integrated Bioinformatics and Machine Learning for Ascertainment and Validation of Biomarkers for Screening Breast Disease. Genes 2025, 16, 1389. https://doi.org/10.3390/genes16111389
Wang Q, Yang S, Zhang Y, Piao C, Liu X, Wu X. Integrated Bioinformatics and Machine Learning for Ascertainment and Validation of Biomarkers for Screening Breast Disease. Genes. 2025; 16(11):1389. https://doi.org/10.3390/genes16111389
Chicago/Turabian StyleWang, Qi, Saisai Yang, Yao Zhang, Chengyu Piao, Xin Liu, and Xiuhong Wu. 2025. "Integrated Bioinformatics and Machine Learning for Ascertainment and Validation of Biomarkers for Screening Breast Disease" Genes 16, no. 11: 1389. https://doi.org/10.3390/genes16111389
APA StyleWang, Q., Yang, S., Zhang, Y., Piao, C., Liu, X., & Wu, X. (2025). Integrated Bioinformatics and Machine Learning for Ascertainment and Validation of Biomarkers for Screening Breast Disease. Genes, 16(11), 1389. https://doi.org/10.3390/genes16111389





