Abstract
Background/Objectives: Esterases are widely used in various industrial fields. This study aimed to isolate and characterize esterase genes from Laceyella sacchari HS49-1, a thermophilic bacterium from a hot spring, which can survive at 45–60 °C and pH 5-10 with robust esterase activity. Methods: A genomic shotgun library was constructed to identify three esterase genes: two in family XII (Est2 and Est7) and one in family VIII (Est1). Sequence analysis revealed significant divergence from other genera. Only Est1 was successfully expressed in Escherichia coli. Its activity, optimal conditions, thermostability, and structure were investigated using p-NP butyrate, temperature/pH assays, heating pre-treatment, and fluorescence quenching. Results: Est1 demonstrated high activity (57.43 ± 0.04 U/mg) towards short-chain p-NP butyrate (C4). Molecular-docking analyses revealed that Est1’s catalytic motif (GXSXG) interacts with various p-NP esters, with binding energy and interaction types varying by acyl chain length. The optimal temperature was 60 °C, and the optimal pH was 8. Est1 exhibited excellent thermostability, retaining 90% of its activity after pre-treatment at 50 °C for 8 h and 69.8% after pre-treatment at 80 °C for the same duration. Fluorescence quenching showed that after 1 h at 80 °C, the fluorescence was reduced by only 16.6%, indicating remarkable heat resistance. Additionally, Est1 did not require metal ions as cofactors and maintained 74.8% of its activity in the presence of 0.1% SDS. Conclusions: The unique properties of Est1 from L. sacchari HS49-1 highlight its potential for industrial applications. Further exploration of this thermophilic bacterium could uncover more valuable genes.
1. Introduction
Lipolytic enzymes play a pivotal role across a spectrum of industrial sectors, including food processing, bioremediation, pharmaceuticals, detergent manufacturing, biodiesel production, and waste management [1]. The market for microbial esterases, especially lipases, was expected to be valued at $684.55 million in2025 and is projected to grow to $963.71 million by 2030 [2]. These enzymes, which are capable of hydrolyzing carboxyl esters of varying acylglycerol chain lengths, are categorized into two primary groups based on their enzymatic activities: triacylglycerol acylhydrolases, commonly referred to as lipases (EC 3.1.1.3), and carboxylic-ester hydrolases, known as esterases (EC 3.1.1.1) [3]. In the classification of lipolytic enzymes, there are 35 distinct families have been recognized. Among these, family I, which encompasses 11 subfamilies, is distinguished as the true lipases, while families II through XXXV are classified as esterases [4]. Microbial esterase, which primarily belong to the α/β-hydrolase family, are characterized by a conserved catalytic triad consisting of serine, histidine, and aspartic acid [5,6]. This structural feature is responsible for their broad substrate specificity and high catalytic efficiency.
Various esterases have been identified from diverse microbial sources. For instance, EstGoM, an autotransporter esterase derived from a marine Pseudomonas sp., exhibited the high hydrolytic activity toward p-nitrophenyl octanoate, with optimal activity at pH 9 and 60 °C [7]. Moreover, EstGtA3, an esterase from the thermophilic bacterium Geobacillus thermodenitrificans NG80-2, preferred short-chain substrates and exhibited peak activity at 60 °C [8]. Esterases derived from hyperthermophiles, such as Thermotoga, Thermoanaerobacter, Aeropyrum, Archaeoglobus, Pyrococcus, and Sulfolobus, demonstrated exceptional thermostability, with optimal temperature ranges for activity spanning from 60 to 100 °C [9]. These enzymes prefer short- and medium-chain p-nitrophenyl (p-NP) esters as substrates. These findings underscore the versatility of microbial esterases in degrading diverse substrates and highlight the potential of these thermostable esterases for industrial applications.
The genus Laceyella currently comprises five validated species (L. sacchari, L. putida, L. sediminis, L. tengchongensis, and L. thermophila) according to the NCBI database [10,11,12,13], which were originally classified under Thermoactinomyces before taxonomic reclassification [14]. These bacteria, which are Gram-positive, thermophilic, spore-forming, and aerobic, belong to the family Thermoactinomycetaceae. Their thermophilic characteristics have prompted extensive studies on various heat-resistant proteins from this genus, demonstrating significant biotechnological potential. Thermitase from L. sacchari DSM 43353 exhibits thermostable endo-protease activity, maintaining stability at 50 °C with functional activity across pH 6–11 [15]. A poly(L-lactide)-degrading serine protease from L. sacchari LP175 shows optimal activity at 60 °C and pH 9.0 [16], while β-1,3-glucanase from L. putida FM3001 displays optimal curdlan hydrolysis at pH 4.2 and 80 °C [17]. Additionally, L-leucine dehydrogenase from L. sacchari catalyzes reversible oxidative deamination/reductive amination of branched-chain L-amino acids with optimal activity at 60 °C [18], and α-amylase from Laceyella sp. DS3 functions optimally at 55 °C and pH 6–7 [19]. These studies collectively demonstrate that proteins derived from this genus maintain remarkable activity and stability under elevated temperatures, highlighting their substantial potential for industrial applications.
In this study, we successfully isolated a thermophilic bacterial strain from the hot spring of Taiwan, which was taxonomically classified as L. sacchari. This strain was deposited in the Bioresource Collection and Research Center (BCRC), Taiwan, with the accession number BCRC AC10098. Genomic analysis of the strain revealed the presence of esterase genes, and we constructed a genomic library to screen for genes encoding highly active esterases. The subsequent heterologous expression of the identified esterase gene in Escherichia coli allowed for the purification and biochemical characterization of the recombinant enzyme, which we designated as Est1. Notably, Est1 exhibited remarkable thermostability, retaining substantial catalytic activity even at extremely high temperatures.
2. Materials and Methods
2.1. Isolation, Cultivation, and Identification of Strain HS49-1
Samples of sediment and water were collected from the Tenguxi hot spring in Yilan County, Taiwan (24.5550684° N, 121.5159281° E). Approximately 100 mL of water and 10 g of sediment were collected in sterile polypropylene tubes, placed in insulated containers to maintain ambient temperature, and processed within 12 h after sampling. To isolate thermophilic actinomycetes, 1 g of sediment (or 1 mL of water sample) was first enriched in 20 mL of yeast extract–malt extract (YM) broth (yeast extract 4.0 g/L, malt extract 10.0 g/L, dextrose 4.0 g/L) and incubated at 45 °C for 24 h under aerobic conditions with gentle shaking. After enrichment, the culture was serially diluted (10−1–10−6) with sterile saline solution (0.85% NaCl), and aliquots were spread onto YM agar plates (YM broth supplemented with 15.0 g/L agar). Plates were incubated at 45 °C for 3–5 days under aerobic conditions. Multiple isolates exhibiting distinct morphology were obtained and screened for esterase activity using 4-nitrophenyl butyrate as the substrate. Among these isolates, strain HS49-1 exhibited the highest esterase activity and was therefore selected for further physiological and biochemical characterization. Optimal cultivation conditions of strain HS49-1 were determined by growing the isolate in YM medium under various temperatures and pH conditions. Enzymatic profiling was performed with the API ZYM kit (bioMérieux, Marcy-l’Étoile, France) at both 37 °C and 55 °C, and reactions were identified visually according to color development. Genomic DNA was extracted from HS49-1, and 16S rDNA was amplified with universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′), followed by Sanger DNA sequencing. A phylogenetic tree was constructed in MEGA X using the neighbor-joining algorithm with 1000 bootstrap replicates to assess node reliability.
2.2. Genomic Sequencing of L. sacchari HS49-1
The genomic sequencing of L. sacchari HS49-1 was conducted by Genomics Bioscience & Technology Co., Ltd. (New Taipei City, Taiwan). DNA sequencing was performed using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). Raw sequencing reads were processed by Trimmomatic to remove adapter sequences and low-quality bases [20]. Subsequently, de novo assembly was carried out using the Shovill pipeline, version 0.9 (https://github.com/tseemann/shovill) (accessed on 31 October 2022). Gene prediction and annotation of the assembled genome were analyzed with Prokka, resulting in multiple predicted protein-coding sequences [21]. Functional annotation of the predicted proteins was performed using COG (Clusters of Orthologous Groups) [22], GO (Gene Ontology) [23], and KEGG (Kyoto Encyclopedia of Genes and Genomes) [24,25] databases to elucidate protein functions and metabolic pathways.
2.3. Construction of a Genomic Shotgun Library from L. sacchari HS49-1
Genomic DNA was cloned using the pEZSeq™ Blunt Cloning Kit (pEZSeq Amp, Lucigen, now Biosearch Technologies, Hoddesdon, UK). DNA was sheared to 1–4 kb fragments with a Hydroshear instrument (BST Scientific, Rockville, MD, USA), size-selected on a 1% agarose gel, and purified by gel extraction. The recovered fragments were quantified with a NanoDrop spectrophotometer (NanoDrop ND-1000, Thermo Fisher Scientific, Inc., Waltham, MA, USA), end-repaired, and ligated into the pEZSeq Amp vector. Ligation products were electroporated into E. coli EPI300™ (Epicentre, now Illumina, Madison, WI, USA) competent cells. Transformed cells were plated on LB agar containing 4 mM IPTG, 40 µg mL−1 X-Gal, 10 µL mL−1 triolein, and 100 µg mL−1 ampicillin and then incubated overnight at 37 °C. Positive clones exhibiting esterase activity (clear hydrolysis zones) were selected for DNA sequencing. These clones were designated TS1, TS2, and TS7. The esterase-encoding genes derived from the TS1, TS2, and TS7 clones were designated Est1, Est2, and Est7, respectively. A phylogenetic tree of the esterases was constructed with the neighbor-joining algorithm implemented in MEGA X, using 1000 bootstrap replicates to assess node reliability.
2.4. Construction of Esterase Expression Plasmids
Gene synthesis and expression vector construction were commissioned to Azenta Life Sciences (Suzhou, China). The genes Est1, Est2, and Est7 were subcloned into the pET15b expression vector without any codon optimization. The pET15b plasmid was first linearized with BamHI. The synthetic genes Est1, Est2, and Est7 were then individually joined to the linearized plasmid using the Gibson assembly method [26]. The 5′ and 3′ ends of the synthetic genes were designed with homologous sequences to the pET15b plasmid, which are CCTGGTGCCGCGCGGCAGCCATATGCTCGA and GCTGCTAACAAAGCCCGAAAGGAAGC, respectively. Each insert gene was in-frame with the vector’s coding sequence. All expression plasmids were verified by full-length DNA sequencing to confirm both sequence accuracy and correct reading frame.
2.5. Overexpression and Identification of Esterases in E. coli
The expression plasmid was introduced into E. coli C43(DE3) competent cells by heat-shock transformation. Transformed cells were plated onto LB agar containing the ampicillin and incubated at 37 °C for 12–16 h until single colonies appeared. A single colony was then transferred to 5 mL LB medium supplemented with ampicillin and cultivated at 37 °C with shaking for 8–12 h to obtain an overnight culture. The starter culture was inoculated into 250 mL fresh LB medium in a 500 mL conical flask and grown at 37 °C with shaking at 175 rpm (THZ-98C, Shanghai Bluepard instruments Co., Ltd., Shanghai, China) for 3–4 h until the OD600 reached 0.4–0.6. Protein expression was induced by adding IPTG to a final concentration of 0.5 mM, and cultivation continued with shaking at 175 rpm (THZ-98C) for 12–16 h at 25 °C. After induction, cells were harvested by centrifugation, the supernatant was discarded, and the pellet was resuspended in Native Buffer (25 mM Tris-HCl, 150 mM NaCl, pH 7.0). The suspension was kept on ice and lysed by sonication. The lysate was clarified by centrifugation, and 10% (v/v) glycerol was added to the supernatant, which was stored at −20 °C until further use. This procedure was performed in triplicate.
After SDS-PAGE, proteins were transferred to a PVDF membrane at 4 °C by wet electroblotting. The membrane was immediately immersed in 20 mL of blocking solution (5% non-fat milk in TBST) and incubated for 1 h at room temperature with gentle shaking. Following blocking, the membrane was incubated overnight at 4 °C with gentle agitation in 4 mL of the same blocking buffer containing anti-6 × His Tag mouse monoclonal antibody (BBI Life Sciences, Shanghai, China) diluted 1:4000 (1 µL antibody stock in 4 mL buffer). The next day, the membrane was washed three times with PBST (5 min per wash). It was then incubated with HRP-conjugated goat anti-mouse IgG secondary antibody (1:4000 in blocking buffer) for 1 h at room temperature with shaking. After another three PBST washes (5 min each), equal volumes of ECL-A and ECL-B substrates from the SuperSignal™ West Pico Kit (Thermo Fisher Scientific, Waltham, MA, USA) were mixed and applied to the membrane. Following a 1-min incubation in the dark, chemiluminescence was captured using a Gel Doc XR+ imaging system (Bio-Rad, Hercules, CA, USA).
2.6. Recombinant Esterase Proteins Purification
Protein purification was performed using a Ni-NTA column according to the manufacturer’s protocol (Ni-NTA Purification System; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The supernatant obtained after sonication was mixed with Ni-NTA resin and incubated for 3 h at 4 °C with gentle agitation to allow His-tagged protein binding. The resin was allowed to settle, and the flow-through was carefully discarded. The column was then washed five times with 15 mL Native Wash Buffer (50 mM NaH2PO4, 0.5 M NaCl, pH 8.0, supplemented with 20 mM imidazole, pH 6.0); after each wash the resin was allowed to sediment and the buffer was drained. Bound protein was eluted with 10 mL Native Elution Buffer (50 mM NaH2PO4, 0.5 M NaCl, pH 8.0, containing 250 mM imidazole, pH 6.0). The eluate was immediately loaded into a Vivaspin® 6 ultrafiltration concentrator (Sartorius AG, Göttingen, Germany), concentrated by centrifugation, and collected. Glycerol was added to a final concentration of 10% (v/v), and the concentrated protein was stored at −20 °C.
2.7. Enzymatic Activity Assay of the Esterase
Esterase activity was assayed spectrophotometrically using p-nitrophenyl (p-NP) esters as substrates. Substrates with different acyl-chain lengths were selected, including short-chain p-NP butyrate (C4) and p-NP valerate (C5); medium-chain p-NP caprylate (C8) and p-NP caprate (C10); and long-chain p-NP laurate (C12), p-NP myristate (C14), and p-NP palmitate (C16). For reactions using C4–C10 substrates (short- and medium-chain), the total reaction volume was 800 μL, consisting of 20 μL of 10 mM p-NP ester, 20 μL of ethanol, 740 μL of 50 mM Tris–HCl (pH 7.0), and 20 μL of recombinant esterase (0.1 mg/mL). For reactions using C12–C16 substrates (long-chain), the total reaction volume was 800 μL, consisting of 20 μL of 10 mM p-NP ester, 20 μL of ethanol, 740 μL of a mixture of 0.1% (w/v) gum arabic, 0.1% (w/v) sodium deoxycholate, and 50 mM Tris–HCl (pH 7.0), and 20 μL recombinant esterase (0.1 mg/mL). Reactions were incubated at the optimal temperature and pH for 15 min and immediately quenched on ice. Absorbance was measured at 405 nm (Molecular Devices, Sunnyvale, CA, USA). Activity corresponding to the highest absorbance was defined as 100%; all other values were expressed relative to this maximum. The unit (U) refers to the amount of esterase required to catalyze the formation of 1 μmol of pNP per minute under the same reaction conditions (temperature, pH, etc.).
2.8. Thermostability of the Esterase
The recombinant esterase was pre-incubated in a water bath set at 50–80 °C for 0–8 h. Immediately after heat treatment, residual activity was determined under the condition: the total reaction volume was 800 μL, containing 20 μL of 10 mM p-NP butyrate (C4), 20 μL of ethanol, 740 μL of 50 mM Tris–HCl (pH 7.0), and 20 μL of pre-heated enzyme (0.1 mg/mL). The reaction was initiated by transferring the mixture to 60 °C for exactly 15 min, then quenched on ice. Absorbance at 405 nm was recorded spectrophotometrically. The highest absorbance obtained at 0 h was defined as 100%, and all other activities were expressed relative to this maximum.
Fluorescence emission spectra of the enzyme were acquired under two thermal regimes. Temperature-gradient protocol: aliquots of recombinant esterase (0.1 mg/mL) were heated for 1 h at 50, 60, 70 or 80 °C, then immediately cooled on ice. Time-gradient protocol: aliquots were pre-incubated at 80 °C for 0, 2, 4, 6 or 8 h before detecting residual enzymatic activities. All samples were transferred to a black 96-well plate and scanned with a microplate reader (excitation: 280 nm, emission: 300–500 nm, slit widths: 4.66 nm for both excitation and emission) (Molecular Devices).
2.9. Effect of Various Reagents on the Esterase
The influence of various chemical factors on esterase activity was assessed spectrophotometrically containing 20 μL of 10 mM p-NP butyrate (C4), 20 μL of ethanol and 740 μL of 50 mM Tris–HCl buffer (pH 7.0), supplemented with 50 mM KCl, 50 mM CaCl2, 50 mM MgCl2, 50 mM NaCl, 0–10 mM EDTA, 0.1% (w/v) SDS or 0.1% (v/v) H2O2, respectively. After enzyme addition, the mixture was incubated at 60 °C for 15 min, immediately quenched on ice, and the absorbance at 405 nm was measured (Molecular Devices). The activity of the additive-free control was set to 100% and the relative activity of each treatment was calculated as the ratio of its absorbance to that of the control.
2.10. Data Statistical Analysis
All experimental data are presented as mean ± standard deviation. All of our experiments were performed in triplicate. Differences among group means were compared using Duncan’s multiple range test, with statistical significance set at a 95% confidence level (p < 0.05). This analysis was specifically conducted to assess the activity of Est1 with a variety of p-nitrophenyl esters as substrates. All statistical analyses were performed using IBM SPSS Statistics software, version 31 (SPSS Inc., Chicago, IL, USA).
2.11. Accession Numbers
The strain L. sacchari HS49-1 has been deposited at the Bioresource Collection and Research Center (BCRC) of the Food Industry Research and Development Institute in Taiwan, with the accession number BCRC AC10098. The whole-genome sequencing data of this strain have been deposited in GenBank under the accession number PRJNA1305130. Additionally, the Est1, Est2, and Est7 genes have been deposited in GenBank under the accession numbers PX121224, PX121225, and PX121226, respectively.
3. Results
3.1. Identification and Characterization of Strain HS49-1
Strain HS49-1 was isolated from a hot spring source. Growth characterization revealed that the thermophilic strain exhibited an optimal growth temperature of 55 °C, with a viable growth range between 45 and 60 °C (Table S1). The strain demonstrated remarkable pH tolerance, growing across a broad pH range of 5 to 10, with optimal growth observed at pH 6–7 (Table S2). Enzyme activity profiling using the API ZYM system showed temperature-dependent enzymatic activities (Table S3). At 37 °C, only esterase (C4), esterase lipase (C8), and naphthol-AS-B1-phosphohydrolase displayed significant activity, while leucine arylamidase and α-chymotrypsin showed weak activity. When the reaction temperature was increased to 55 °C, multiple enzymes exhibited pronounced activity, including alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, α-chymotrypsin, naphthol-AS-B1-phosphohydrolase, and α-glucosidase. Phylogenetic analysis based on 16S rRNA gene sequencing and NCBI database comparisons revealed that HS49-1 formed a distinct cluster with L. sacchari, L. sediminis, and L. tengchongensis, showing closest similarity to L. sacchari (Figure 1). This strain has been deposited in the Bioresource Collection and Research Center (Food Industry Research and Development Institute, Taiwan) under the designation L. sacchari BCRC AC10098.
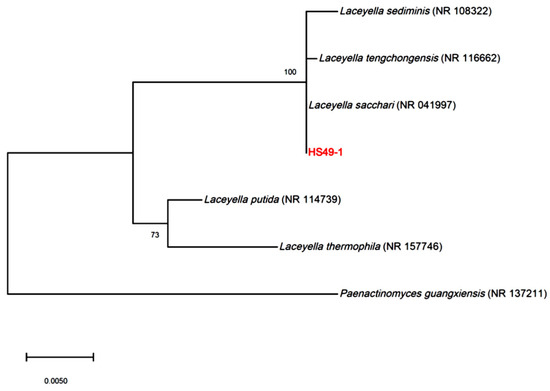
Figure 1.
Phylogenetic tree based on 16S rDNA sequences, showing the evolutionary relationships of strain HS49-1 with species of the genus Laceyella, including L. sediminis (NR 108322), L. tengchongensis (NR 116662), L. sacchari (NR 041997), L. putida (NR 114739), and L. thermophila (NR 157746). Paenactinomyces guangxiensis (NR 137211) is included as an outgroup for comparative analysis to root the tree. Bootstrap values (based on 1000 replicates) are shown at the nodes, indicating the reliability of branching.
3.2. Draft Genome Sequence of L. sacchari HS49-1
The genome of L. sacchari HS49-1 comprised 3,269,339 bp, which is relatively smaller among the compared Laceyella species (Table S4). Notably, L. putida possessed the largest genome at 4,075,529 bp. Strain HS49-1 contained 3350 predicted genes, a number that is moderate and commensurate with its genome size among the compared strains. The GC content of HS49-1 was determined to be 49%, which was comparable to other Laceyella species. Moreover, comparative analysis identified distinct differences in esterase and lipase gene content, with HS49-1 exhibiting a significantly higher number of annotated esterase genes. Functional annotation through GO analysis demonstrated enrichment in DNA-binding and oxidoreductase activities (molecular function), transmembrane transport and ribosome-related components (cellular component), and transmembrane transport processes (biological process) (Figure S1). COG classification revealed the most abundant functional categories to be general function prediction, followed by amino acid transport and metabolism, and inorganic ion transport and metabolism. KEGG pathway analysis indicated the strain’s involvement in multiple amino acid biosynthesis and degradation pathways (including alanine, phenylalanine, and tryptophan metabolism), carbohydrate biosynthesis and degradation, as well as key energy metabolic pathways.
3.3. Esterase Activity Screening Using Genomic Shotgun Library
Chromosomal DNA from L. sacchari HS49-1 was sheared to generate 1–4 kb fragments and used to construct a genomic shotgun library. Transformants were plated on LB agar containing 1% triolein and screened for esterase activity based on clear-zone formation. Three clones—TS1, TS2 and TS7—displayed pronounced esterase activity (Figure 2). Notably, the TS1 clone produced the largest clear zone, indicating the strongest esterase activity among the three positive isolates. These three clones were selected for further DNA sequencing analysis. The results revealed open reading frames encoding proteins of 284, 523 and 537 amino acids, designated Est1, Est2 and Est7, respectively. Phylogenetic comparison with characterized esterase families placed Est1 in family VIII, while Est2 and Est7 clustered within family XII (Figure 3).
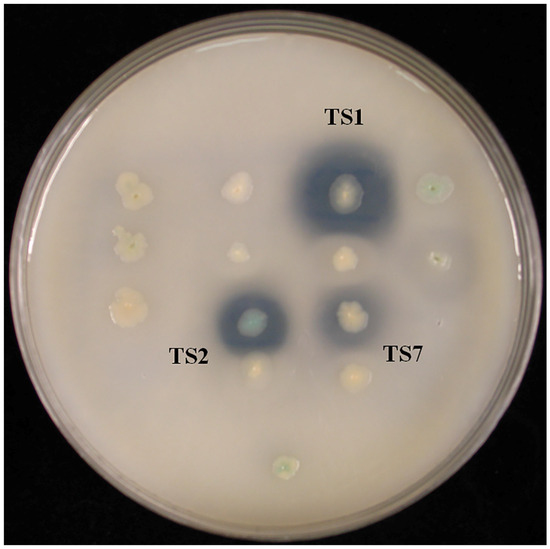
Figure 2.
Transformants derived from the genomic shotgun library of L. sacchari HS49-1 were plated on LB agar supplemented with 1% triolein to assess esterase activity via clear-zone formation.
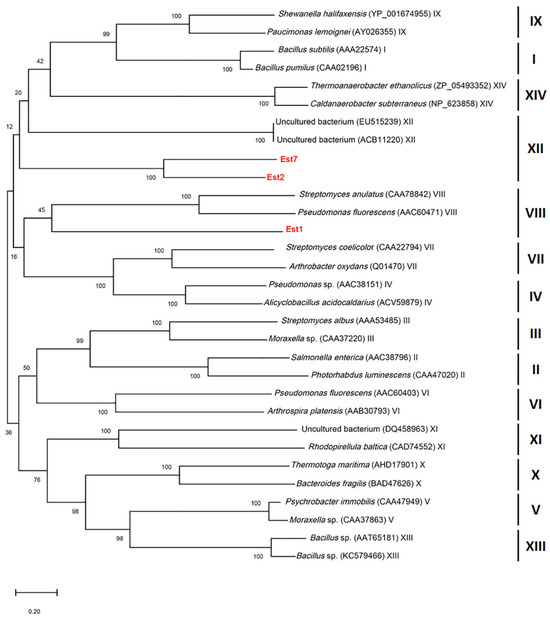
Figure 3.
Phylogenetic tree constructed based on esterase sequences, showing the evolutionary relationships of Est1, Est2, and Est7 with esterases from various bacterial taxa and uncultured bacteria. Bootstrap values (based on 1000 replicates) are shown at the nodes, indicating the reliability of branching.
3.4. Construction and Overexpression of Recombinant Esterases in E. coli
To purify the three esterases, the corresponding genes were sub-cloned into the pET15b vector to yield N-terminal His-tagged constructs in E. coli. IPTG induction was used to drive expression, and the presence of each protein was verified by Western blotting with an anti-His antibody. SDS-PAGE analysis of whole-cell lysates revealed no discernible overexpression bands regardless of IPTG (Figure 4). However, Western blotting demonstrated a strong immunoreactive band exclusively for Est1 after IPTG induction; Est2 and Est7 remained undetectable. Consequently, Est1 was purified by Ni2+-NTA affinity chromatography, yielding a single dominant band on SDS-PAGE that matched the predicted molecular mass of 31.8 kDa (Figure 5). The purification process, utilizing a conical flask in conjunction with sonication and Ni2+-NTA affinity chromatography, achieved a yield of Est1 protein of 0.47 ± 0.09 mg/L.
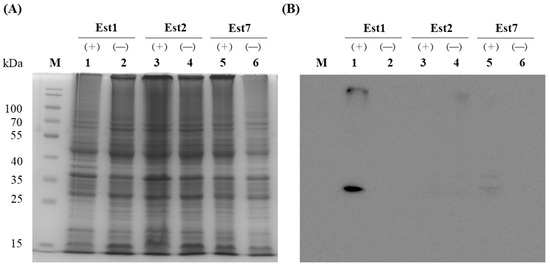
Figure 4.
Overexpression of recombinant Est1, Est2, and Est7 in E. coli C43(DE3). (A) Coomassie blue-stained SDS-PAGE gel showing esterase proteins from soluble cell-free extracts, with lanes 1, 3, and 5 representing samples induced with 0.5 mM IPTG (+) and lanes 2, 4, and 6 representing uninduced controls (−). (B) Western blot analysis of esterase proteins from soluble cell-free extracts, with lanes 1, 3, and 5 corresponding to 0.5 mM IPTG-induced samples (+) and lanes 2, 4, and 6 to uninduced controls (−). M indicates the protein marker.
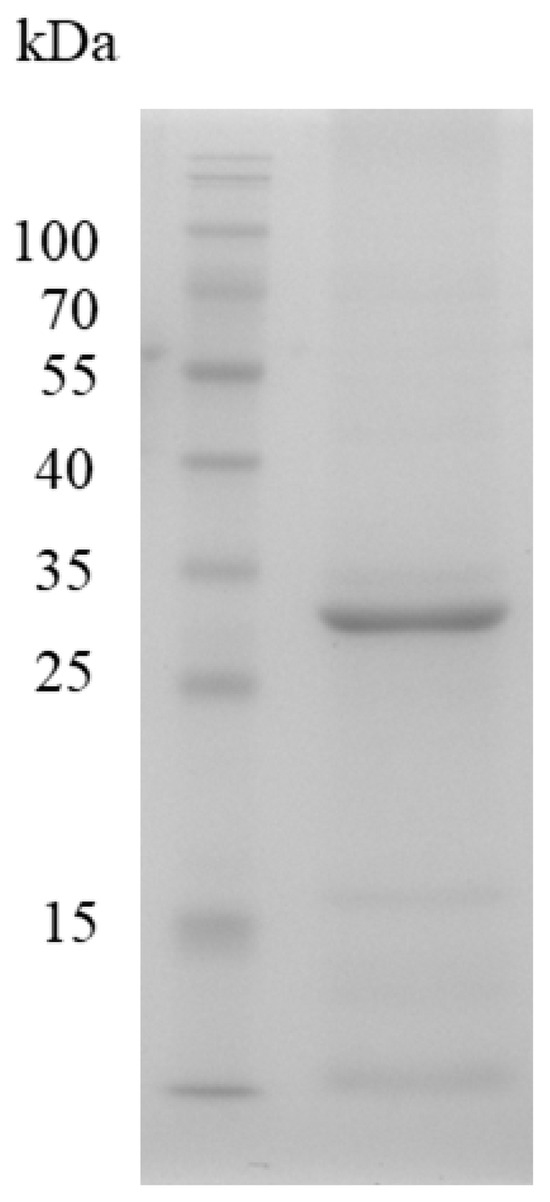
Figure 5.
SDS-PAGE analysis of Est1 protein purification. A prominent band on the gel, observed after purification via a nickel column, indicates the Est1 protein. M indicates the protein marker.
3.5. Characterization of the Recombinant Esterase, Est1
Using p-nitrophenyl (p-NP) esters as substrates, esterase activity was determined spectrophotometrically. The catalytic profile of recombinant Est1 was then systematically investigated with a homologous series of p-NP esters: p-NP butyrate (C4), p-NP valerate (C5), p-NP caprylate (C8), p-NP caprate (C10), p-NP laurate (C12), p-NP myristate (C14), and p-NP palmitate (C16). Recombinant Est1 displayed robust hydrolytic activity toward short-chain substrates—namely p-NP butyrate (C4), p-NP valerate (C5), and p-NP caprylate (C8)—with a marked preference for p-NP butyrate (C4) (Figure 6). Based on the analysis of enzyme activity, the results indicate that Est1 exhibited activities of 57.43 ± 0.04, 13.00 ± 0.15, and 10.08 ± 0.26 U/mg towards p-NP butyrate (C4), p-NP valerate (C5), and p-NP octanoate (C8), respectively.
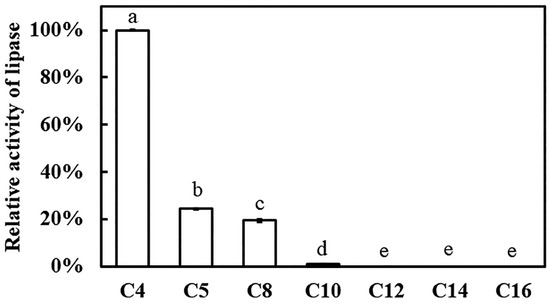
Figure 6.
Evaluation of Est1 activity using various p-nitrophenyl esters as substrates. Relative activity was calculated with the highest activity set as 100%. Reactions were performed at pH 7.0 and 60 °C for 15 min, followed by analysis of absorbance at 405 nm. Different letters indicate significant differences among groups.
To rationalize these findings, molecular-docking analyses were performed between recombinant Est1 and each p-NP ester (Figure 7). The three-dimensional structure of Est1 was predicted using SWISS-MODEL homology modeling [27]. The conserved GXSXG catalytic motif has also been identified in the Est1 [28], and all these various substrates were indeed found to interact with Est1’s catalytic motif (specifically at S152) [29]. As the acyl chain lengthened, the calculated binding energy progressively decreased, accompanied by an increased number of hydrophobic contacts (Table 1). Notably, the longest substrates, p-NP myristate (C14) and p-NP palmitate (C16), established additional π–π stacking interactions. In contrast, the shortest substrate, p-NP butyrate (C4), formed only one π-cation interaction, five hydrogen bonds, and four hydrophobic interactions with the enzyme, underscoring the distinct binding mode.
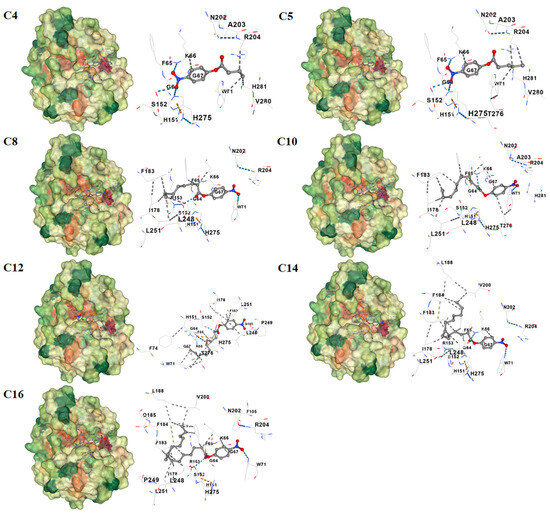
Figure 7.
Molecular docking analysis of Est1 with various p-nitrophenyl esters. The three-dimensional structure of Est1 was predicted via SWISS-MODEL homology modeling, and molecular docking was performed using CD-Dock2 (https://cadd.labshare.cn/cb-dock2/index.php) (accessed on 25 June 2025). The dashed lines in blue, grey, orange, and green correspond to hydrogen bonds, hydrophobic interactions, π–cation interactions, and π–π stacking, respectively.

Table 1.
Molecular docking simulations were conducted using CB-Dock2 to systematically assess the binding interactions between Est1 and a series of substrates with varying acyl-chain lengths (C4-C16).
Due to the thermophile L. sacchari HS49-1, recombinant Est1 was profiled across a broad temperature and pH spectrum to define its optimal catalytic activity (Figure 8). The enzyme peaked at 60 °C yet exhibited extraordinary thermostability. The decline in activity beyond the optimum was gradual, retaining 85% of its maximal rate at 100 °C. Remarkably, even at 10 °C recombinant Est1 preserved 53.5% relative activity, underscoring its broad thermal adaptability. In contrast to this pronounced temperature tolerance, recombinant Est1 displayed a markedly narrower pH range. Activity was maximal at pH 8.0, whereas values below 6.0 or above 8.0 led to almost complete loss of function, revealing stringent requirements for mildly alkaline conditions.
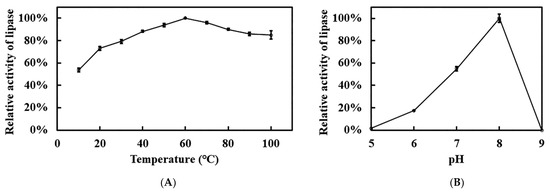
Figure 8.
Optimal temperature and pH analyses of Est1. (A) Temperature profile of Est1, determined at 10–100 °C under pH 7 for 15 min. (B) pH profile of Est1, assessed at 60 °C using 50 mM sodium acetate/acetic acid buffer (pH 5), 100 mM sodium phosphate buffer (pH 6–8), and glycine/NaOH buffer (pH 9) for 15 min.
3.6. Thermostable Assay of the Recombinant Esterase, Est1
To assess the long-term thermostability of recombinant Est1, aliquots were pre-incubated at 50–80 °C for up to 8 h and residual activity was subsequently determined. After 8 h of pre-incubation at 50 °C, the enzyme retained 90% of its initial activity (Figure 9). As the pre-incubation temperature rose, a gradual decline in stability was observed. Relative activities of 83.6%, 80.2%, and 69.8% remained after 8 h at 60 °C, 70 °C, and 80 °C, respectively. Notably, within the 50–80 °C, recombinant Est1 maintained over 90% relative activity for at least the first 2 h, underscoring its robust thermal stability and suitability for high-temperature applications.
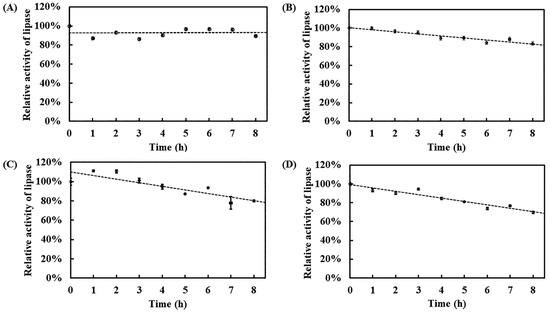
Figure 9.
Thermostability analysis of Est1 at various temperatures. Est1 was incubated at (A) 50 °C, (B) 60 °C, (C) 70 °C, and (D) 80 °C for 8 h. Residual enzyme activity was measured at 60 °C for 15 min, followed by absorbance analysis at 405 nm. The circles represent the residual activity of Est1 after being subjected to various temperatures for different durations. The line illustrates the trend over time following these treatments.
According to the intrinsic fluorescence of tryptophan, tyrosine, and phenylalanine, we examined how temperature and incubation time affected the tertiary structure of recombinant Est1. The native enzyme exhibited an average maximum emission wavelength of 328–331 nm (Figure 10). Pre-incubation at 50 °C or 60 °C for 1 h left the fluorescence spectrum unchanged, indicating preserved conformation. Raising the temperature to 70 °C or 80 °C reduced the fluorescence intensity by only 12.1% and 16.6%, respectively, revealing modest short-term perturbations. To assess long-term thermal stress, recombinant Est1 was pre-treated at 80 °C for 0–8 h. Fluorescence measurements revealed a gradual decline in intensity, ultimately reaching a 53% reduction after 8 h of observation. This pronounced quenching demonstrates that prolonged high-temperature exposure substantially disrupts the protein structure.
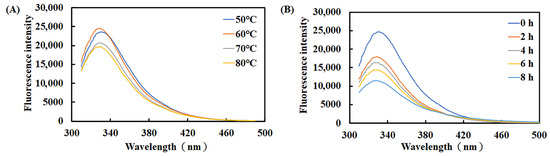
Figure 10.
Fluorescence spectra of Est1 following treatment at different temperatures and durations. (A) Est1 was incubated at 50–80 °C for 1 h. (B) Est1 was incubated at 80 °C for 8 h. Fluorescence spectra were recorded using a microplate reader.
3.7. Impact of Chemical Reagents on the Recombinant Esterase, Est1, Activity
The influence of various chemical reagents and metal ions on recombinant Est1 activity was showed in Figure 11. Among the additives tested, H2O2 exerted the strongest inhibitory effect, followed by the anionic surfactant SDS. Among the metal ions, Ca2+ produced a notable change in activity. To clarify whether Est1 requires a metal cofactor, the enzymatic activity with increasing concentrations of EDTA was performed. Even at 10 mM EDTA, activity remained essentially unchanged, indicating that Est1 catalysis proceeds without the assistance of bound metal ions.
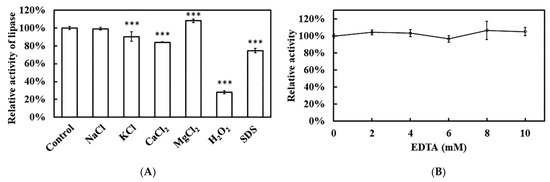
Figure 11.
Effect of various reagents on Est1 activity. (A) Different reagents (50 mM NaCl, KCl, CaCl2, MgCl2; 0.1% H2O2, and SDS) and (B) EDTA (0–10 mM) were added to assess Est1 activity at 60 °C for 15 min. Residual enzyme activity was determined via absorbance analysis at 405 nm. *** indicates a significant difference compared with the control group.
4. Discussion
The isolated strain, based on 16S rDNA sequencing and genomic data, revealed high similarity to L. sacchari. Comparative studies showed L. sacchari HS49-1 had a relatively narrow growth temperature range (45–60 °C) compared to other Laceyella type strains: L. sacchari KCTC 9790T (35–65 °C), L. putida KCTC 3666T (30–65 °C), L. sediminis RHA1T (28–65 °C), L. tengchongensis YIM 10002T (28–70 °C), and L. thermophila YIM 79486T (37–65 °C) [10,11,12,13]. Like other members of this genus, it tolerated a broad pH range while preferring neutral conditions for optimal growth [10,11,12]. API ZYM assays detected significant esterase activity in L. sacchari HS49-1 at 37 °C, with additional enzymatic activities such as alkaline phosphatase and α-glucosidase emerging at 55 °C, indicating the presence of thermostable enzymes. As no systematic studies of Laceyella esterases have been reported previously, this study employed genome mining to identify and characterize novel esterases in this thermophilic genus.
In this study, we first constructed a genomic shotgun library of L. sacchari HS49-1 and conducted plate-based screening for esterase activity. Three distinct clones were identified, each forming clear zones on triolein plates—an indication of lipolytic activity. The three esterase gene clones (Est1, Est2, and Est7) from L. sacchari HS49-1 demonstrated high sequence conservation within the genus, with α/β hydrolase gene sequences showing 71.4–100% similarity among different Laceyella species (L. tengchongensis, L. sacchari, and L. putida) according to the NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 8 July 2025). In contrast, when compared to esterases from other genera within the same family Thermoactinomycetaceae, including Thermoactinomyces (WP_170151265.1), Salinithrix (WP_380701314.1), and Paenactinomyces (WP_181752047.1), the sequence similarity dropped significantly to 51.32–64.52%. This substantial reduction in similarity highlighted the unique characteristics of Laceyella’s esterase genes, suggesting they have evolved distinct structural or functional features that differentiate them from related genera in the Thermoactinomycetaceae family.
Nevertheless, when these genes were subcloned into pET15b expression vectors driven by the T7 promoter, only the family VIII esterase, Est1, was successfully expressed. While factors such as codon optimization, fusion tags, vector selection, and promoter strength are known to influence protein expression [30], our analysis revealed no obvious inclusion body formation, suggesting the problem likely occurs during the translation process. These results implies that although the T7 promoter system is highly effective for recombinant protein production, it may be unsuitable for certain genes—particularly family XII esterase genes, which appear sensitive to the strong transcriptional activity of T7 promoters. This sensitivity could create bottlenecks during transcription or translation [31]. Additionally, the strong transcriptional activity driven by T7 promoters may lead to more complex mRNA secondary structures, particularly in the translation initiation region. These intricate structural conformations could potentially impede ribosomal binding and subsequent assembly, thereby significantly compromising translation initiation efficiency [32].
Analysis of Est1 activity toward p-nitrophenyl substrates with varying chain lengths demonstrated that Est1 exhibited optimal catalytic efficiency for the short-chain p-NP butyrate (C4), with activity progressively declining as the carbon chain length increases. Molecular docking studies revealed that while all substrates form essential hydrogen bonds with the enzyme, longer-chain substrates develop increasingly extensive hydrophobic interactions with Est1’s binding pockets. These findings indicate that substrate binding was stabilized through a combination of hydrogen bonding and hydrophobic interactions, both crucial for forming the enzyme-substrate complex and facilitating catalysis [33]. However, the enhanced hydrophobic interactions observed with longer-chain substrates, although strengthening binding affinity, appear to hinder product release, thereby reducing overall catalytic efficiency [34]. This trade-off between binding stability and catalytic turnover explained the inverse correlation observed between substrate chain length and enzymatic activity, highlighting the delicate balance required between substrate binding and product release in enzymatic catalysis.
Thermostable esterases often exhibit increased hydrophobicity in their protein sequences [35]. This elevated proportion of hydrophobic amino acids reinforces the protein’s hydrophobic core, thereby improving structural stability at high temperatures. Additional stabilizing factors include the formation of salt bridges (e.g., between Asp-Lys or Glu-Arg pairs) [36], an increased presence of secondary structural elements (α-helices and β-sheets) [37,38], and the formation of disulfide bonds, which collectively mitigate thermal denaturation [39]. Furthermore, specific residues such as proline, contribute to stability due to the rigidity of their side chains [40]. To investigate these mechanisms, we compared several Family VIII esterases, including PBS-2 (Paenibacillus sp.) [41], Est13L, DLFae4, and EstCS3 (metagenome-derived) [42,43,44], with optimal activity temperatures of 30 °C, 40 °C, 50 °C, and 55 °C, respectively. Notably, the thermostability of Est1 (60 °C) surpassed all these homologs (Table S5). Secondary structure prediction via YASPIN revealed that Est1 did not exhibit the highest number of α-helices or β-strands [45], suggesting that its thermostability is not primarily driven by these features. Instead, Est1 displayed a significantly higher proline content. Comparative analysis of hydrophobic (AILFWV) and charged (RKHYCDE) amino acids highlighted that Est1 and DLFae4 possessed the highest hydrophobic content, with phenylalanine, and valine being major contributors. These findings suggest that thermostability in Family VIII esterases is predominantly achieved through enrichment of hydrophobic residues, with Est1 serving as a striking example of this trend.
The relative activity of the esterase under various treatments revealed distinct responses to ions, oxidizing agents, and detergents. Notably, NaCl, KCl, CaCl2, and MgCl2 had minimal impact on enzyme activity, with values remaining close to the control. This suggests that the esterase was highly tolerant to monovalent (Na+, K+) and divalent (Ca2+, Mg2+) cations, which may stabilize its structure or had negligible interference with the active site. In contrast, H2O2 and SDS significantly reduced activity, with SDS showing a more pronounced inhibitory effect. The sensitivity to H2O2 implies that oxidative modification (e.g., of catalytic residues) may disrupt function [46], while the strong inhibition by SDS likely reflects protein denaturation due to disruption of hydrophobic interactions critical for stability [47]. Critically, EDTA had no inhibitory effect across the tested range (0–10 mM), as activity remained unchanged compared to the control. This confirms that the esterase does not require metal cofactors for catalysis or structural stability, distinguishing it from metalloenzymes [48]. The absence of EDTA sensitivity aligns with its tolerance to ionic treatments, reinforcing its metal-independent mechanism.
5. Conclusions
We have identified and characterized a novel esterase, named Est1 and classified under family VIII, originating from the thermophilic bacterium L. sacchari HS49-1—a relatively understudied strain. Genomic analysis revealed significant divergence between the genes of this bacterium and those of its congeners, underscoring the distinctiveness of this genus. Our successful heterologous expression of Est1 in E. coli yielded an enzyme with outstanding thermostability, maintaining 69.8% of its activity after an 8-h incubation at 80 °C. Furthermore, Est1 functioned independently of metal ion cofactors, making its activity less susceptible to interference from metal ions in industrial applications. The remarkable thermal resilience of Est1 highlighted the unique adaptive features of Laceyella-derived enzymes and positioned it as a promising candidate for industrial processes requiring robust biocatalysts under extreme temperatures.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16111330/s1, Figure S1: Genomic analysis of L. sacchari H49-1; Table S1: The growth characteristics of strain HS49-1 were observed by culturing it in YM medium at various temperatures; Table S2: The growth characteristics of strain HS49-1 were observed by culturing it in YM medium at various pH levels; Table S3: The enzymatic activities of strain HS49-1 were analyzed using the API ZYM system at both 37 °C and 55 °C; Table S4: Genome sequence summary of L. sacchari H49-1 and published Laceyella species; Table S5: A comparison of optimal temperature, pH, protein secondary structure, and amino acid sequence composition between Est1 and other proteins belonging to the same family VIII esterases.
Author Contributions
Conceptualization, Y.-P.C. and L.-L.L.; validation, Y.-P.C. and Y.-C.K.; formal analysis, X.Z., H.-J.P., C.-Y.T., M.T., H.W. and F.C.; investigation, Y.-P.C., X.Z., H.-J.P., C.-Y.T., M.T., L.-L.L., H.W., F.C. and Y.-C.K.; resources, Y.-P.C., L.-L.L. and Y.-C.K.; data curation, Y.-P.C., X.Z., H.-J.P. and Y.-C.K.; writing—original draft preparation, Y.-P.C. and H.-J.P.; writing—review and editing, Y.-P.C., X.Z., H.-J.P., C.-Y.T., M.T., L.-L.L., H.W., F.C. and Y.-C.K.; supervision, Y.-P.C.; project administration, Y.-C.K.; funding acquisition, Y.-C.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Student Innovation and Entrepreneurship Training Program of Xiamen Medical College, Fujian, China, grant number X202212631005.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zan, X.; Li, A.; Xu, Y.; Meng, Q.; Luo, T.; Sun, L.; Sun, W.; Cui, F. Lipases from bacteria and fungi: Classification, subcellular localization, and regulation of activity. J. Agric. Food Chem. 2025, 73, 16697–16714. [Google Scholar] [CrossRef]
- Lipase Market Size & Share Analysis—Growth Trends and Forecast (2025–2030). Available online: https://www.mordorintelligence.com/industry-reports/lipase-market (accessed on 26 October 2025).
- Sandoval, G.; Herrera-López, E.J. Lipase, phospholipase, and esterase biosensors (Review). Methods Mol. Biol. 2018, 1835, 391–425. [Google Scholar] [PubMed]
- Hitch, T.C.A.; Clavel, T. A proposed update for the classification and description of bacterial lipolytic enzymes. PeerJ 2019, 7, e7249. [Google Scholar] [CrossRef]
- Zan, X.; Cui, F.; Sun, J.; Zhou, S.; Song, Y. Novel dual-functional enzyme Lip10 catalyzes lipase and acyltransferase activities in the oleaginous fungus Mucor circinelloides. J. Agric. Food Chem. 2019, 67, 13176–13184. [Google Scholar] [CrossRef]
- Yasutake, Y.; Konishi, K.; Muramatsu, S.; Yoshida, K.; Aburatani, S.; Sakasegawa, S.I.; Tamura, T. Bacterial triacylglycerol lipase is a potential cholesterol esterase: Identification of a key determinant for sterol-binding specificity. Int. J. Biol. Macromol. 2021, 167, 578–586. [Google Scholar] [CrossRef]
- Rodríguez-Mejía, J.L.; Hidalgo-Manzano, I.A.; Muriel-Millán, L.F.; Rivera-Gomez, N.; Sahonero-Canavesi, D.X.; Castillo, E.; Pardo-López, L. A novel thermo-alkaline stable GDSL/SGNH esterase with broad substrate specificity from a deep-sea Pseudomonas sp. Mar. Biotechnol. 2024, 26, 447–459. [Google Scholar]
- Curci, N.; Strazzulli, A.; De Lise, F.; Iacono, R.; Maurelli, L.; Dal Piaz, F.; Cobucci-Ponzano, B.; Moracci, M. Identification of a novel esterase from the thermophilic bacterium Geobacillus thermodenitrificans NG80-2. Extremophiles 2019, 23, 407–419. [Google Scholar] [CrossRef]
- Levisson, M.; van der Oost, J.; Kengen, S.W. Carboxylic ester hydrolases from hyperthermophiles. Extremophiles 2009, 13, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Ming, H.; Ji, W.L.; Li, S.; Zhao, Z.L.; Zhang, L.Y.; Meng, X.L.; Zhou, E.M.; Nie, G.X.; Li, W.J. Laceyella thermophila sp. nov., a thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2017, 67, 2953–2958. [Google Scholar] [CrossRef]
- Chen, J.J.; Lin, L.B.; Zhang, L.L.; Zhang, J.; Tang, S.K.; Wei, Y.L.; Li, W.J. Laceyella sediminis sp. nov., a thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2012, 62, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, S.K.; Zhang, Y.Q.; Yu, L.Y.; Klenk, H.P.; Li, W.J. Laceyella tengchongensis sp. nov., a thermophile isolated from soil of a volcano. Int. J. Syst. Evol. Microbiol. 2010, 60, 2226–2230. [Google Scholar] [CrossRef]
- Li, D.; Huang, W.; Wang, C.; Qiu, S. The complete genome sequence of the thermophilic bacterium Laceyella sacchari FBKL4.010 reveals the basis for tetramethylpyrazine biosynthesis in Moutai-flavor Daqu. MicrobiologyOpen 2019, 8, e922. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kim, I.G.; Shin, Y.K.; Park, Y.H. Proposal of the genus Thermoactinomyces sensu stricto and three new genera, Laceyella, Thermoflavimicrobium and Seinonella, on the basis of phenotypic, phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2005, 55, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, C.M.; Madsen, S.M.; Vrang, A.; Hansen, O.C.; Johnsen, M.G. Recombinant expression of Laceyella sacchari thermitase in Lactococcus lactis. Protein Expr. Purif. 2013, 92, 148–155. [Google Scholar] [CrossRef]
- Hanphakphoom, S.; Maneewong, N.; Sukkhum, S.; Tokuyama, S.; Kitpreechavanich, V. Characterization of poly(L-lactide)-degrading enzyme produced by thermophilic filamentous bacteria Laceyella sacchari LP175. J. Gen. Appl. Microbiol. 2014, 60, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Uchimura, K.; Kubota, T.; Nunoura, T.; Deguchi, S. Biochemical and genetic characterization of β-1,3 glucanase from a deep subseafloor Laceyella putida. Appl. Microbiol. Biotechnol. 2016, 100, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Li, Y.; Jia, H.; Wei, P.; Zhou, H.; Jiang, M. Expression, purification and characterization of a thermostable leucine dehydrogenase from the halophilic thermophile Laceyella sacchari. Biotechnol. Lett. 2016, 38, 855–861. [Google Scholar] [CrossRef]
- El-Sayed, A.K.A.; Abou-Dobara, M.I.; El-Fallal, A.A.; Omar, N.F. Heterologous expression, purification, immobilization and characterization of recombinant α-amylase AmyLa from Laceyella sp. DS3. Int. J. Biol. Macromol. 2019, 132, 1274–1281. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Vera Alvarez, R.; Karamycheva, S.; Makarova, K.S.; Wolf, Y.I.; Landsman, D.; Koonin, E.V. COG database update 2024. Nucleic Acids Res. 2025, 53, D356–D363. [Google Scholar] [CrossRef] [PubMed]
- Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; Hill, D.P.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.-Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: Improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- Alias, F.L.; Nezhad, N.G.; Normi, Y.M.; Ali, M.S.M.; Budiman, C.; Leow, T.C. Recent advances in overexpression of functional recombinant lipases. Mol. Biotechnol. 2023, 65, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, T.; Ohtake, K.; Senda, N.; Kiga, D. correlation between in vitro and in vivo gene-expression strengths is dependent on bottleneck process. New Gener. Comput. 2024, 42, 271–281. [Google Scholar] [CrossRef]
- Behloul, N.; Wei, W.; Baha, S.; Liu, Z.; Wen, J.; Meng, J. Effects of mRNA secondary structure on the expression of HEV ORF2 proteins in Escherichia coli. Microb. Cell Fact. 2017, 16, 200. [Google Scholar] [CrossRef]
- Yang, B.; Pu, M.; Khan, H.M.; Friedman, L.; Reuter, N.; Roberts, M.F.; Gershenson, A. Quantifying transient interactions between Bacillus phosphatidylinositol-specific phospholipase-C and phosphatidylcholine-rich vesicles. J. Am. Chem. Soc. 2015, 137, 14–17. [Google Scholar] [CrossRef]
- Yu, Q.; Fan, L.; Duan, Z. Five individual polyphenols as tyrosinase inhibitors: Inhibitory activity, synergistic effect, action mechanism, and molecular docking. Food Chem. 2019, 297, 124910. [Google Scholar] [CrossRef]
- Cheng, W.; Nian, B. Computer-aided lipase engineering for improving their stability and activity in the food industry: State of the art. Molecules 2023, 28, 5848. [Google Scholar] [CrossRef]
- Sürmeli, Y.; Durmuş, N.; Şanlı-Mohamed, G. Exploring the structural insights of thermostable Geobacillus esterases by computational characterization. ACS Omega 2024, 9, 32931–32941. [Google Scholar] [CrossRef]
- Tomazini, A.; Carvalho, M.; Murakami, M.T.; Viviani, V.R. Effect of pH on the secondary structure and thermostability of beetle luciferases: Structural origin of pH-insensitivity. Photochem. Photobiol. Sci. 2023, 22, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Zhu, L.; Zhu, P.; Li, Q. Lysine-based site-directed mutagenesis increased rigid β-sheet structure and thermostability of mesophilic 1,3-1,4-β-glucanase. J. Agric. Food Chem. 2015, 63, 5249–5256. [Google Scholar] [CrossRef]
- Navone, L.; Vogl, T.; Luangthongkam, P.; Blinco, J.A.; Luna-Flores, C.H.; Chen, X.; von Hellens, J.; Mahler, S.; Speight, R. Disulfide bond engineering of AppA phytase for increased thermostability requires co-expression of protein disulfide isomerase in Pichia pastoris. Biotechnol. Biofuels 2021, 14, 80. [Google Scholar] [CrossRef]
- Tang, Y.; Lin, Z.; Ren, L.; Feng, C.; Wang, Y.; Wang, F. Enhancing the thermostability of phospholipase C by structural-based proline incorporation to improve its degumming performance. Appl. Biochem. Biotechnol. 2025, 197, 4837–4849. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Park, I.S.; Nam, B.H.; Kim, D.G.; Jee, Y.J.; Lee, S.J.; An, C.M. A novel esterase from Paenibacillus sp. PBS-2 is a new member of the β-lactamase belonging to the family VIII lipases/esterases. J. Microbiol. Biotechnol. 2014, 24, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- Nan, F.; Jiang, J.; Wu, S.; Zhang, Y.; Qiu, J.; Qiao, B.; Li, S.; Xin, Z. A novel VIII carboxylesterase with high hydrolytic activity against ampicillin from a soil metagenomic library. Mol. Biotechnol. 2019, 61, 892–904. [Google Scholar] [CrossRef]
- Park, J.M.; Won, S.M.; Kang, C.H.; Park, S.; Yoon, J.H. Characterization of a novel carboxylesterase belonging to family VIII hydrolyzing β-lactam antibiotics from a compost metagenomic library. Int. J. Biol. Macromol. 2020, 164, 4650–4661. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Jeong, G.S.; Lee, H.W.; Kim, H. Molecular characterization of novel family IV and VIII esterases from a compost metagenomic library. Microorganisms 2021, 9, 1614. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Simossis, V.A.; Taylor, W.R.; Heringa, J. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 2005, 21, 152–159. [Google Scholar] [CrossRef]
- Gottfredsen, R.H.; Larsen, U.G.; Enghild, J.J.; Petersen, S.V. Hydrogen peroxide induce modifications of human extracellular superoxide dismutase that results in enzyme inhibition. Redox Biol. 2013, 1, 24–31. [Google Scholar] [CrossRef]
- Rasmussen, H.; Wollenberg, D.T.W.; Wang, H.; Andersen, K.K.; Oliveira, C.L.P.; Jørgensen, C.I.; Jørgensen, T.J.D.; Otzen, D.E.; Pedersen, J.S. The changing face of SDS denaturation: Complexes of Thermomyces lanuginosus lipase with SDS at pH 4.0, 6.0 and 8.0. J. Colloid Interface Sci. 2022, 614, 214–232. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, V.; Dubey, V.K.; Srivastava, A.; Garg, S.K.; Singh, V.P.; Arora, P.K. Industrial applications of fungal lipases: A review. Front. Microbiol. 2023, 14, 1142536. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).