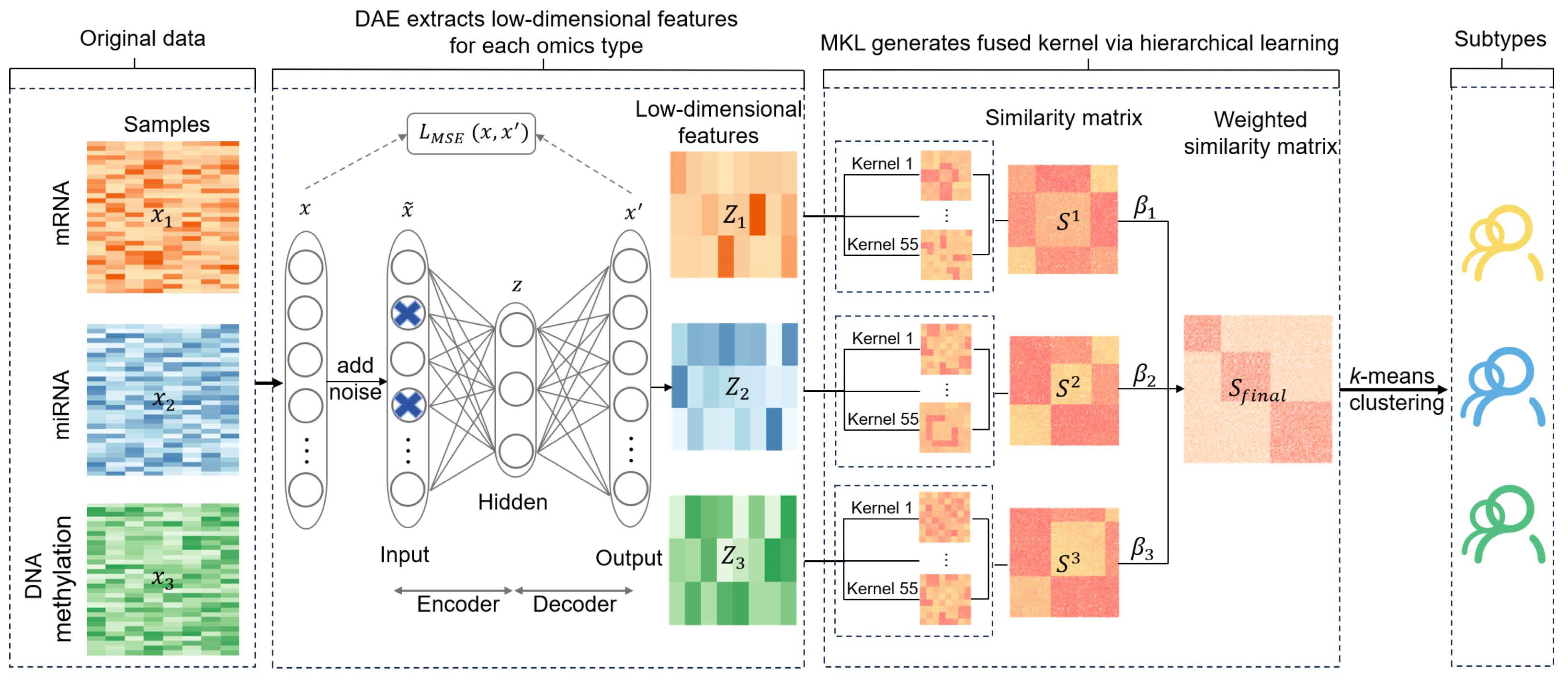
2.1. DAE-MKL
DAE-MKL incorporates a denoising autoencoder within the MKL framework to perform deep-learning dimension-reduction, reconstructing features from multi-omics data to improve subtype identification. It consists of three main steps (See
Figure 1): (1) Deep-learning dimension reduction for each omics data type. DAE is applied to perform nonlinear transformation and feature learning, enabling the extraction of a more robust latent representation for features. (2) Hierarchical multi-kernel learning. The reconstructed features from step (1) serve as input for multi-kernel learning to optimize kernel parameters, where kernel parameters and kernel weight are optimized to learn a composite kernel. These individual kernels are then fused into a final kernel through a weighted linear combination using an unsupervised multiple kernel learning method (UMKL). (3) Cancer subtyping. K-means clustering is performed on the final fused kernel to obtain sample clusters for cancer subtyping.
Stage 1: Extracting low-dimensional representations of each data type using DAE. We first utilize DAE to generate a low-dimensional representation for each omics data type, such as mRNA, miRNA, and DNA methylation. Suppose we have
types of omics data for
patients, which denoted as
. For each omic data type, given input data
, DAE trains the input features with added noise to prevent the network from learning an identity mapping, which would be pointless. This strategy enables the model to better extract meaningful features from noisy data. Common types of noise include Gaussian noise and random dropout:
where
represents Gaussian noise zero mean and a variance
,
randomly drops part of the input. In our study, we adopted random dropout as the noise type for all omics datasets. The noise data
are then used as the input to the encoder. The encoder formula of the autoencoder is:
The hidden layer represents a low-dimensional mapping of the noisy data by the encoder, and then the decoder reconstructs the input data to closely match the original data. This process aims to learn key input features, and the decoder formula is:
Here, the DAE uses the ReLU activation function, defined as
, which is applied to all layers in the encoder and the intermediate layers of the decoder. The goal is to minimize reconstruction error, ensuring that the decoder’s output closely matches the original data. The loss function is introduced to minimize the error between
and
, as follows:
When the loss function is minimized and the number of hidden layer nodes is less than the input layer nodes, the optimal latent feature representation
is obtained. L1 and L2 regularization are then added to enhance the generalization ability and robustness of the autoencoder. After repeated training, the optimal parameters are determined, with the optimal number of hidden layer nodes selected through grid search based on data analysis [
19,
20]. In this study, final hyperparameters, derived from extensive experiments, are as follows: the input and hidden dropout ratios are set to 0.1 and 0.3, respectively, and L1 and L2 regularization values are set to 0.006 and 0.1. A sensitivity analysis exploring different dropout combinations was performed using simulated data (variance = 12, signal-to-noise ratio = 10%), with detailed results presented in
Supplementary Note S1 (Table S1 and Figure S1). The analysis indicates that optimal clustering performance is achieved with input dropout 0.1–0.3 and hidden dropout 0.3–0.5. The resulting low-dimensional feature matrices are denoted as
, with dimension
.
Stage 2: Obtain the fused kernel under the hMKL framework. Given the refined representations
for each data type from stage 1, a hierarchical kernel learning strategy, hMKL, is employed, which first learns a composite kernel by optimizing the kernel parameters and weights for each data type, and then integrates all composite kernels into a final kernel under the UMKL framework.
Step 1. Construct Gaussian kernels and optimize their parameters and weights within the CIMLR framework to learn sample–sample similarities for each omics data type.
The Gaussian kernel [
13] function is defined as follows:
where
is the Euclidean distance between sample
and
. The variance can be calculated as:
where
represents samples that are top
neighbors of the samples
. A total of 55 Gaussian kernels can be can be constructed for each data type by varying the parameters
, where
with a step size of 0.25, and
with a step size of 2 (See Ramazzotti et al. [
14] for details). The distance and kernel between samples
and
can generally be represented as:
, where
represents the weight of each kernel
.
represents the importance of each individual kernel
.
We compute the pairwise similarities between samples across multiple data types through the following optimization framework under the CIMLR method:
where
represents the matrix trace, and
and
are two non-negative tuning parameters,
and
denote the identity matrices of size
and
, respectively, with
is the predefined number of classes,
denotes the Frobenius norm of the similarity matrix
, and
is a low-dimensional matrix that imposes a low-rank structure on
. The optimization objective involves solving three variables: the similarity matrix
, the weight vector
, and the low-rank constraint matrix
. The final similarity matrix
for each data type is then obtained.
Step 2. Obtain the final-weighted similarity matrices within the UMKL framework.
Based on the similarity matrix
of each data type from step 1, UMKL first constructs a
k-nearest neighbor graph
, which is associated with each kernel. Then, an
matrix
that represents the original topological structure of the data, where
indicates the frequency with which the pair
appears in the edge list of
. Specifically, an N-dimensional vector
is introduced, defined as
, where
represent the similarities between sample
and the other samples. UMKL directly uses the kernel matrix
to measure the topological structure between samples, rather than using distances in the original data space. The optimization problem is as follows:
where
is the
th similarity matrix,
is the total number of similarity matrix. Equation (8) can be rewritten as:
where
can be obtained by solving the Quadratic Programming problem in Equation (9) and
for each omics is derived by solving the
constraint in the QP problem. This allows for the obtained of the weights for each omics similarity matrix, representing the relative contribution of each omics. The final fused kernel matrix is expressed as:
where
represents the weights of
.
Stage 3: Use the
k-means clustering to identify cancer subtypes. Based on the final fused kernel matrix
,
k-means clustering method [
21] is applied to obtain sample clusters. Assuming
k-means divides all samples into
clusters, denoted as
, the objective is to minimize the squared error
:
where
is the mean of
.
2.3. Simulation Study
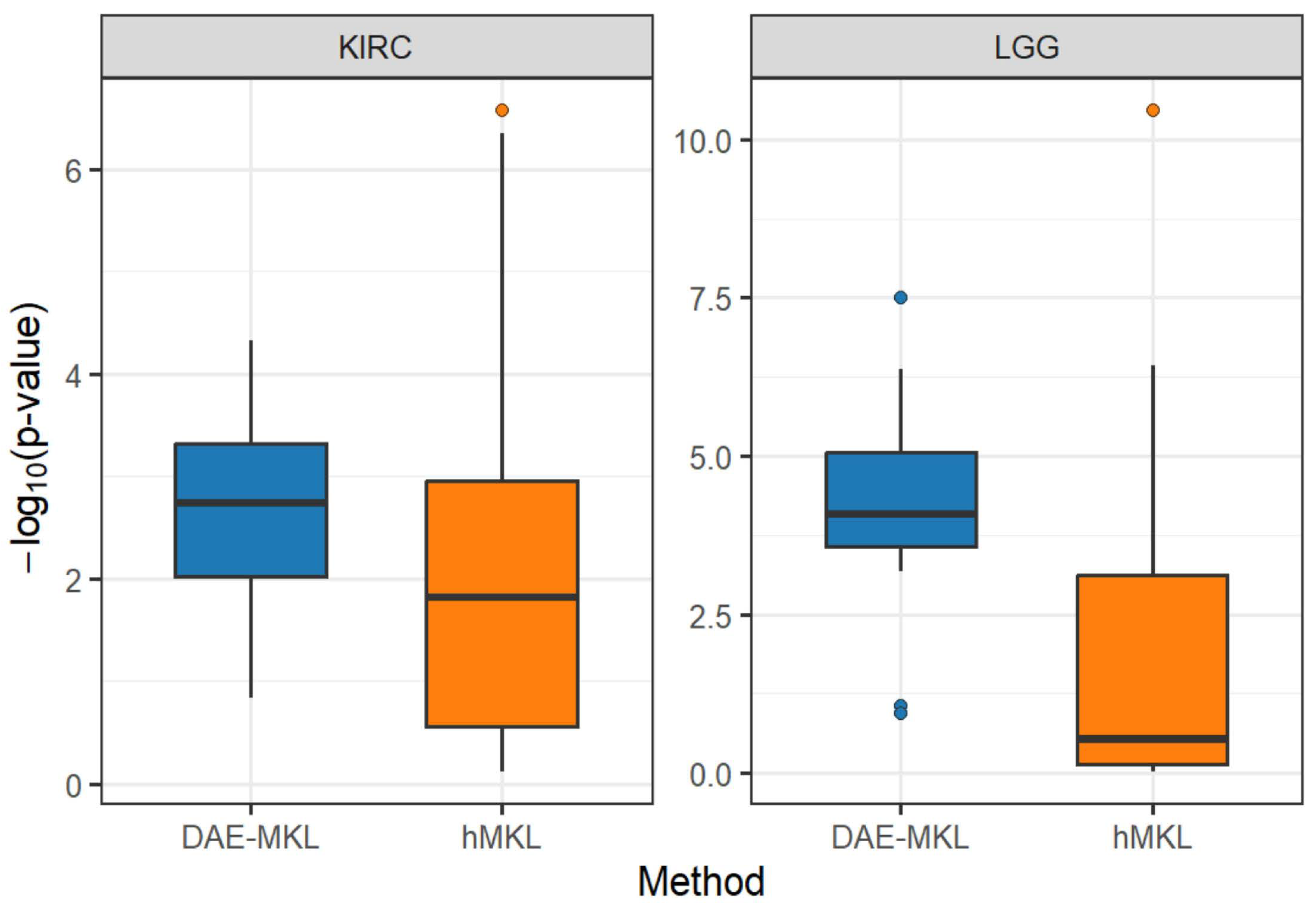
We conducted simulation studies to evaluate the performance of the DAE-MKL method for subtype identification using multi-omics data and compared it with several state-of-the-art methods, including SAE-MKL, AE-MKL, hMKL, CIMLR [
14] and SNF [
13]. Similar to DAE-MKL, the SAE-MKL and AE-MKL approaches are constructed upon sparse autoencoders (SAE) and autoencoders (AE) architectures, respectively, and integrate multi-kernel learning for subtype identification. The simulation design follows the procedures outlined in the literature [
20,
22,
23], with four subtype groups being simulated across three omics data types. The overall clustering structure could only be obtained by integrating information from all omics types, which cannot be achieved at a single omics level. Here, two simulation scenarios were considered. In Scenarios I, we simulated three types of omics data, each consisting of 200 samples and 1000 features. These 200 samples were pre-defined into four subtypes, with each subtype containing 50 samples. Integrating all three omics data is essential for accurately classifying the four subtypes. Considering the independence and overlap among the datasets, three datasets were constructed using the formula
, where
represents the mean expression level of features for each dataset, and
represents random Gaussian noise. In Scenarios Ⅱ, we combined real data from the Gene Expression Omnibus (GEO) database with predefined cluster structures. The GEO database includes GSE10645 [
24] for RNA expression, GSE73002 [
25] for miRNA expression and GSE51557 [
26] for DNA methylation. Singular value decomposition (SVD) was applied to decompose and reconstruct the actual genomic data with predefined cluster structures. In the two simulation scenarios described above, three levels of noise were set with variances of 4, 8, and 12, respectively, and corresponding signal ratios of 5%, 7.5%, and 10%.
In each scenario, we constructed two datasets: SimData1 and SimData2. SimData1 has a clear boundary between subtypes, whereas SimData2 possesses fuzzy boundaries. SimData2 is based on SimData1, which involves randomly sampling 10–20% of the samples in each subtype.
2.5. Multi-Omics Data and Data Processing
We focused on subtypes of LOWER-grade gliomas (LGG) and Kidney Renal Clear Cell Carcinoma (KIRC). LGG, classified as World Health Organization (WHO) grades II and III based on their histopathological features, are among the most common infiltrative tumors in the adult cerebral hemispheres. Some of these neoplasms, which are rarely curable, may progress to transform into higher-grade tumors (WHO grade IV, Glioma) [
28,
29]. Due to the insufficient availability of reliable biomarkers for accurately predicting overall survival in LGG patients, combined with the significant heterogeneity within this tumor group, further molecular subtyping has become increasingly essential. KIRC is one of the most prevalent forms of renal cell carcinoma (RCC), accounting for approximately 70–80% of RCC cases. This subtype is notably aggressive, frequently leading to metastasis and poor prognosis [
30]. The heterogeneity among patients makes it difficult to replicate individualized treatment plans. Therefore, conducting extensive research into the molecular mechanisms of KIRC and developing strategies to guide personalized treatment remains essential [
31].
The Glioma dataset (including mRNA expression, miRNA expression, DNA methylation, and clinical data) was obtained from the CGGA database, while the KIRC dataset was downloaded from the TCGA website using the TCGAbiolinks software (version 2.36.0) [
32]. For LGG, we selected samples classified as pathologic grades II and III to serve as the LGG dataset. The LGG and KIRC datasets were preprocessed as follows:
(1) For LGG datasets, we first selected samples that contained all three omics data, while excluding samples with missing values in clinical factors such as overall survival, survival status, age, gender, and WHO grade, and obtained 86 LGG samples with 827 miRNAs, 19,416 mRNAs, and 14,476 methylation genes. The publicly available multi-omics data were pre-normalized and quality-controlled by the data providers, ensuring consistency and comparability across samples. Next, we performed feature selection based on the most variant Median Absolute Deviation (MAD) to select the top number of value features.
(2) For KIRC datasets, annotated the promoter region CpG methylation sites within 2 kbp of the transcription start site [
33], and removed the CpG sites located on sex chromosomes. The remaining CpG sites were mapped to genes, and the mean beta value of multiple CpG sites per gene was used as the gene-level methylation signal; Features with a deletion ratio greater than 30% were removed, and the remaining missing values were imputed using the K-nearest neighbor (KNN) algorithm [
34]. Meanwhile miRNA and mRNA data were transformed using a
conversion. After these steps, we obtained 285 KIRC samples with 388 miRNAs, 16,893 mRNAs, and 14,296 methylation genes. Subsequently, we performed feature selection based on the most variant Median Absolute Deviation (MAD) to select the top number of value features for each type of omics data.
After preprocessing, we obtained 86 LGG samples with 500 miRNAs, 15,000 mRNAs, and 10,000 methylation genes, as well as 285 KIRC samples with 388 miRNAs, 15,000 mRNAs, and 10,000 methylation genes.
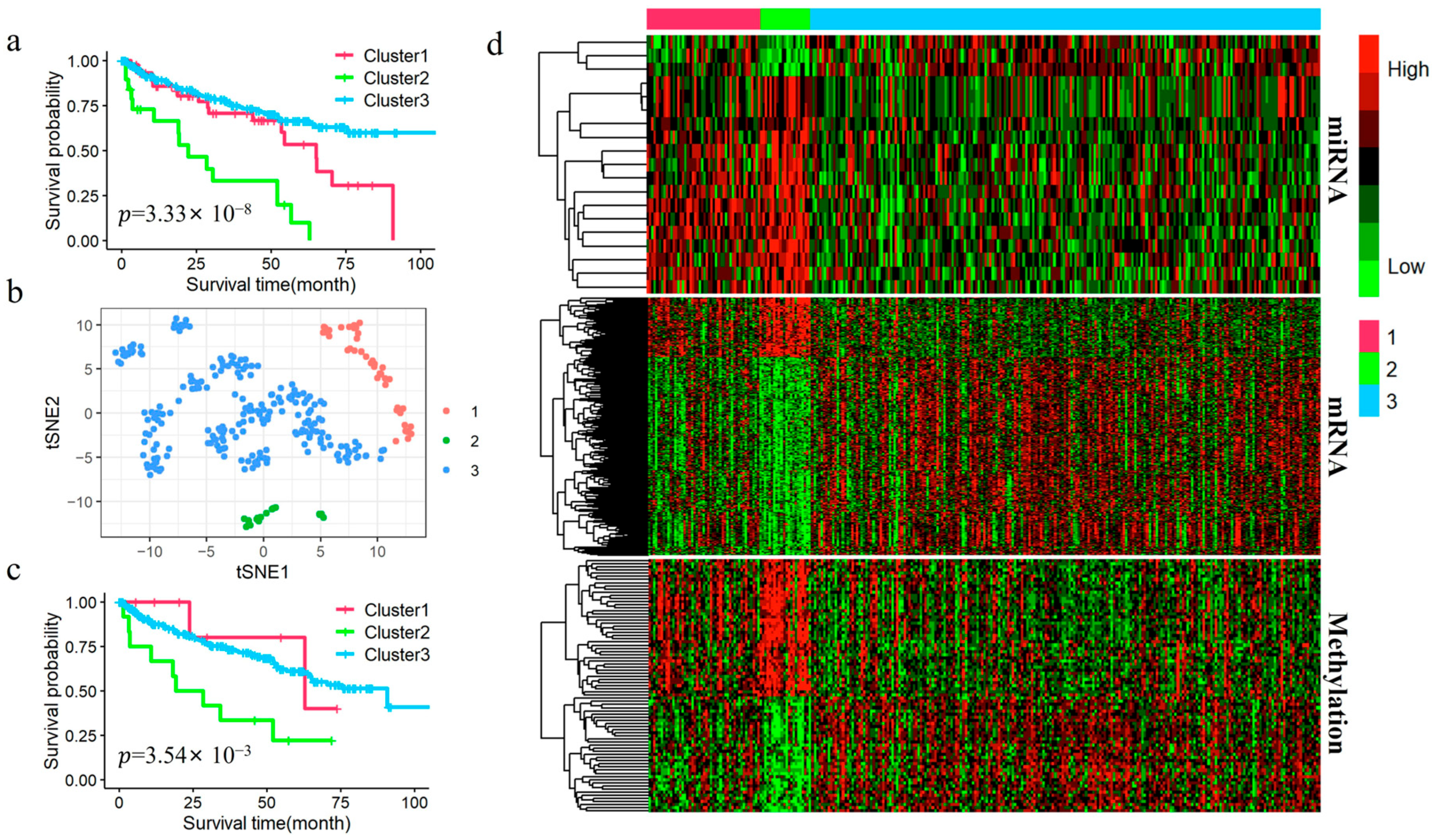
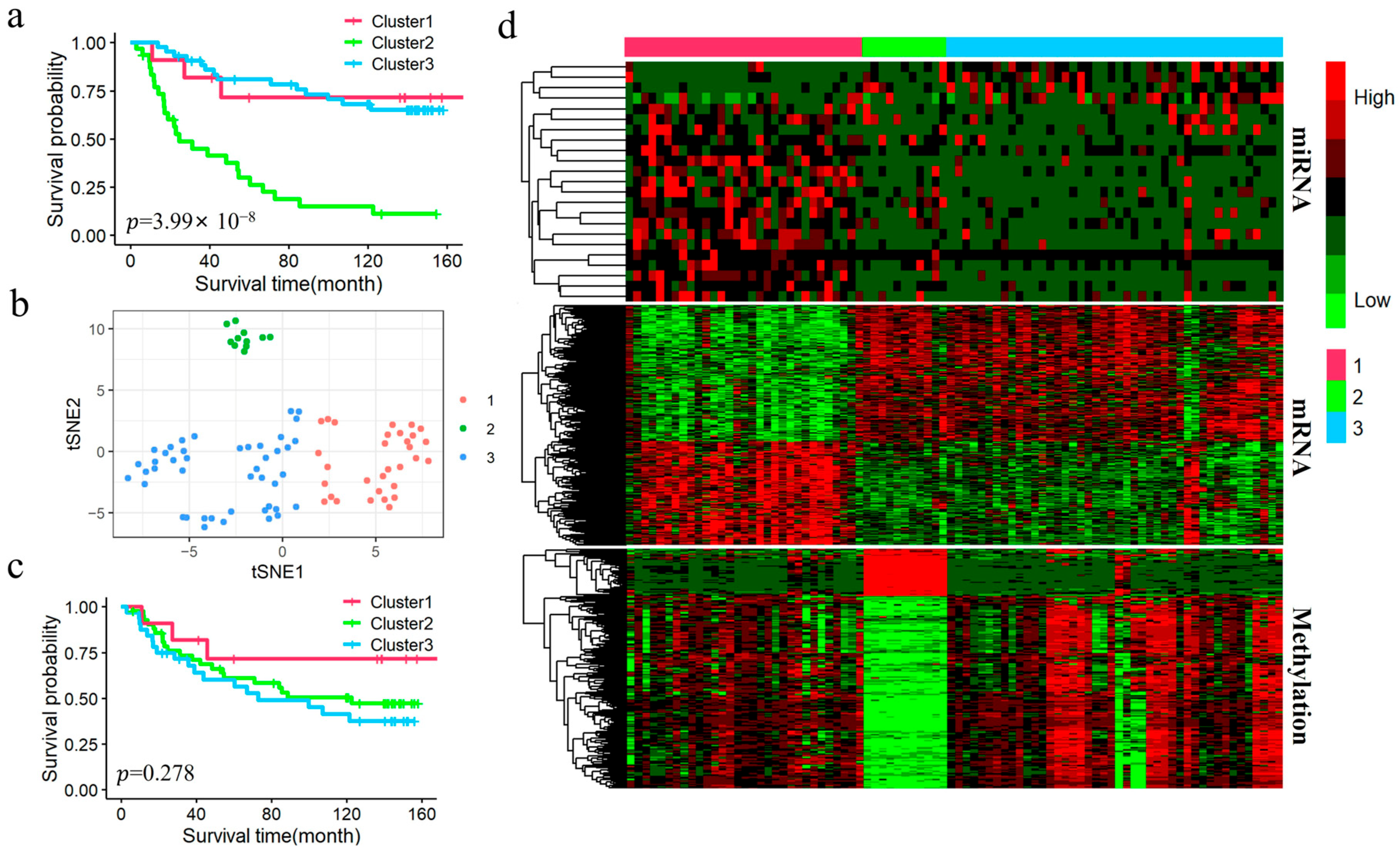
2.6. Downstream Statistical Analysis After Subtyping
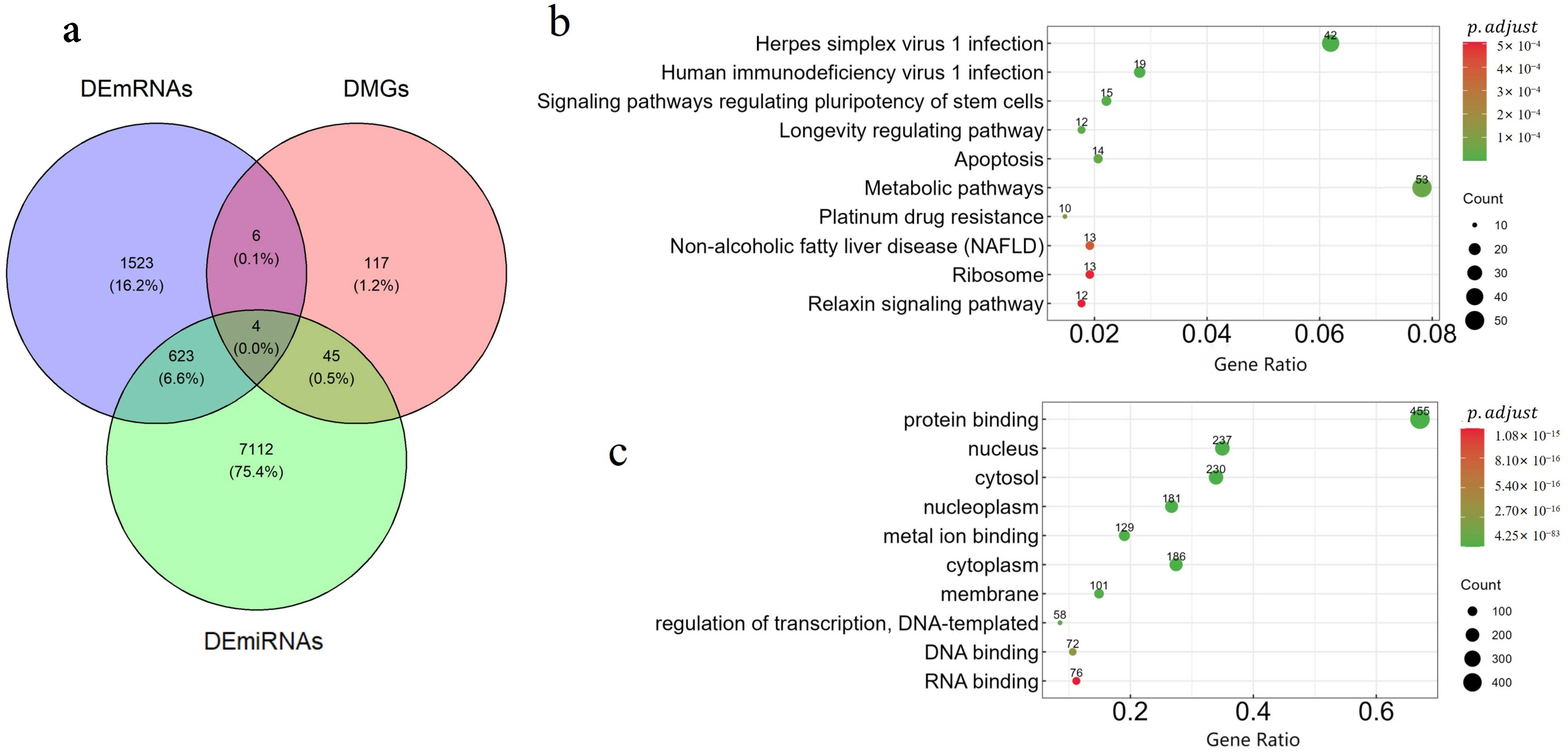
Differential analysis: We performed differential expression analysis to explore the molecular heterogeneity and further validate the biological significance of each subtype. The Kruskal–Wallis H test was used to identify differentially expressed miRNAs (DEmiRNAs), differentially expressed mRNAs (DEmRNAs), and differentially methylated genes (DMGs), with a significance threshold set at an FDR-adjusted
p-value < 0.05. To further assess feature enrichment in each subtype, the hypergeometric distribution test [
14] was employed, applying a filtering criterion of
< 0.05. Additionally, miRWalk [
35] was utilized to predict the target genes of the identified DEmiRNAs.
KEGG and GO enrichment analysis: Gene enrichment analysis was conducted to elucidate the biological processes and pathways represented in the omics data, thereby providing deeper insights into the underlying molecular mechanisms [
36]. Well-established methods for enrichment analysis include the Kyoto Encyclopedia of Genes and Genomes (KEGG) [
37] and Gene Ontology (GO) [
38] analyses. KEGG enrichment analysis focuses on understanding the functional roles of genes and the pathways they are involved in, while GO enrichment analysis categorizes the functions of differentially expressed genes into three main groups: Biological Process (BP), Molecular Function (MF), and Cellular Component (CC). We utilized the R package clusterProfiler (version 4.16.0) [
39] to conduct (GO) and (KEGG) enrichment analyses on the overlapping genes across the three omics datasets.
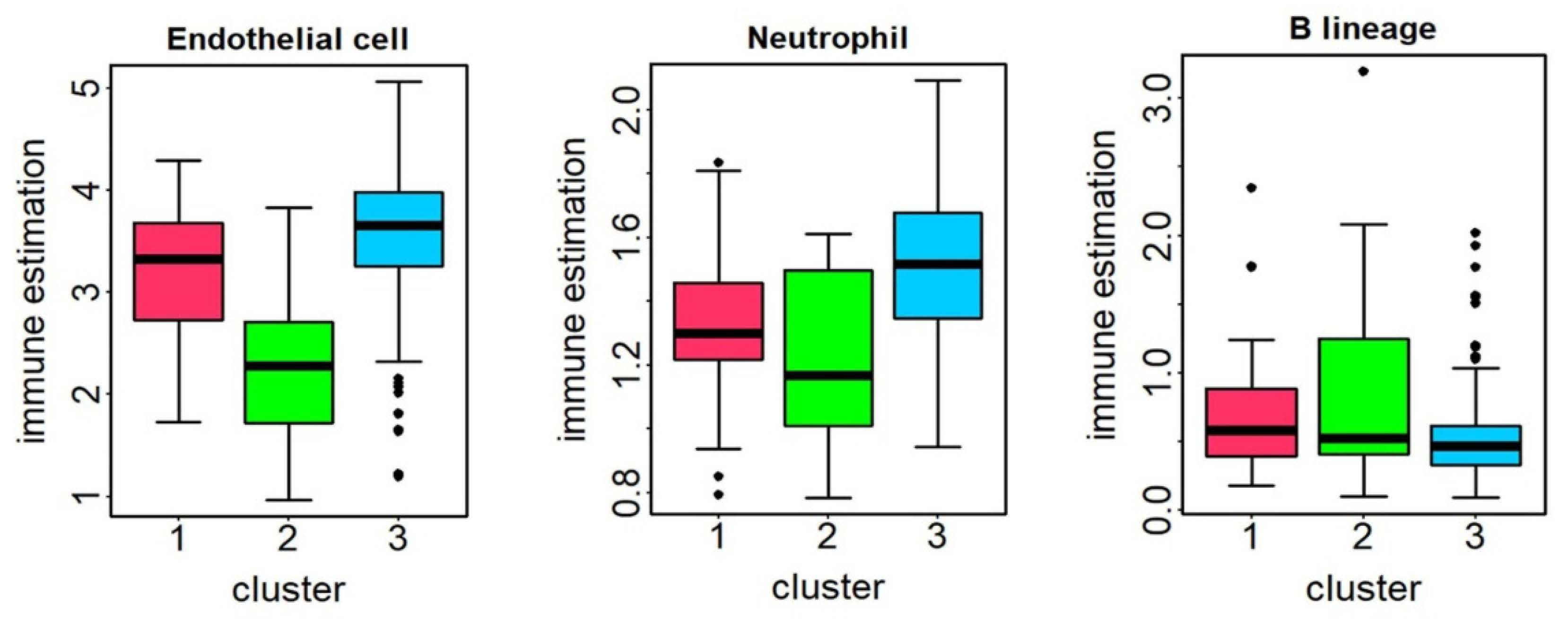
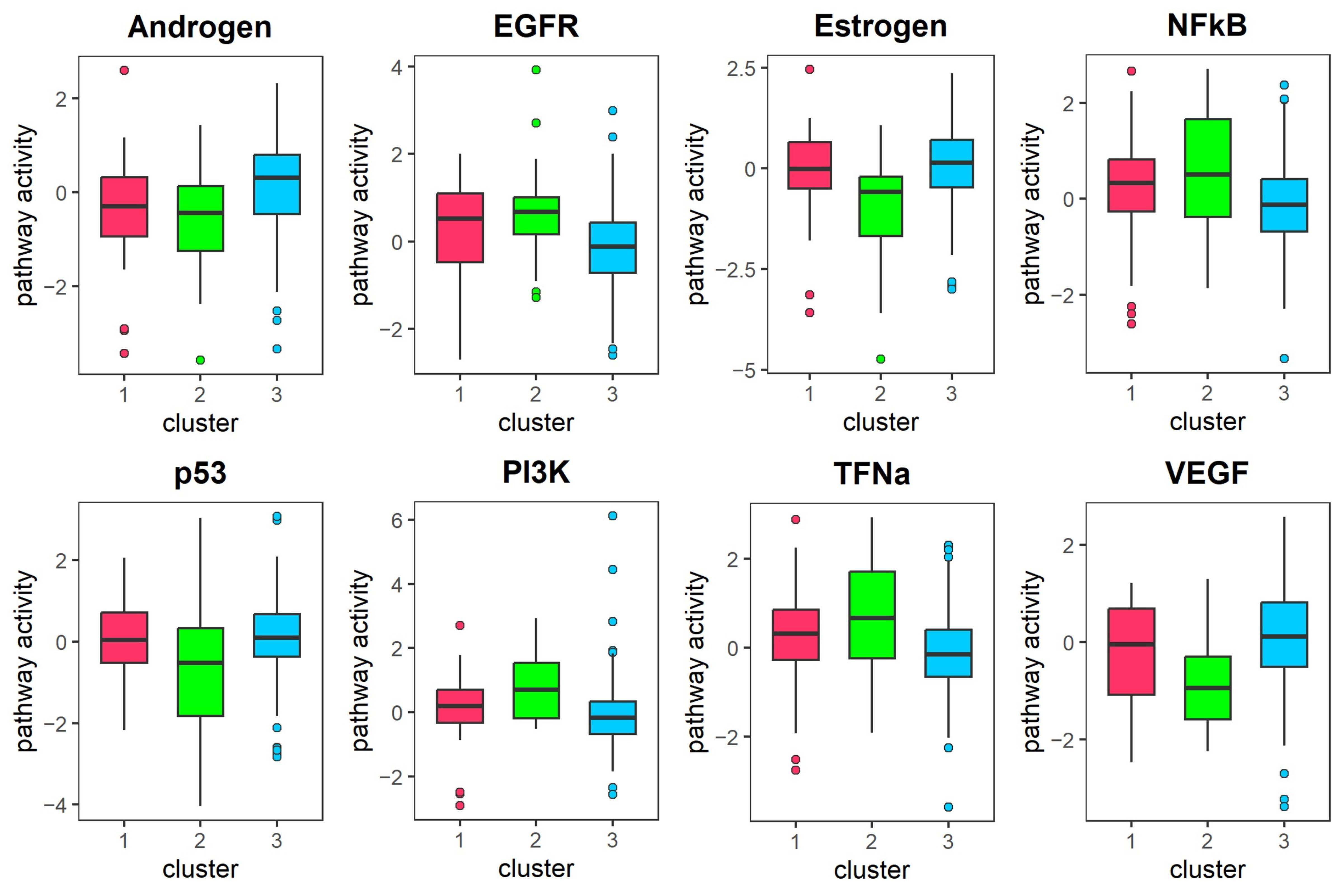
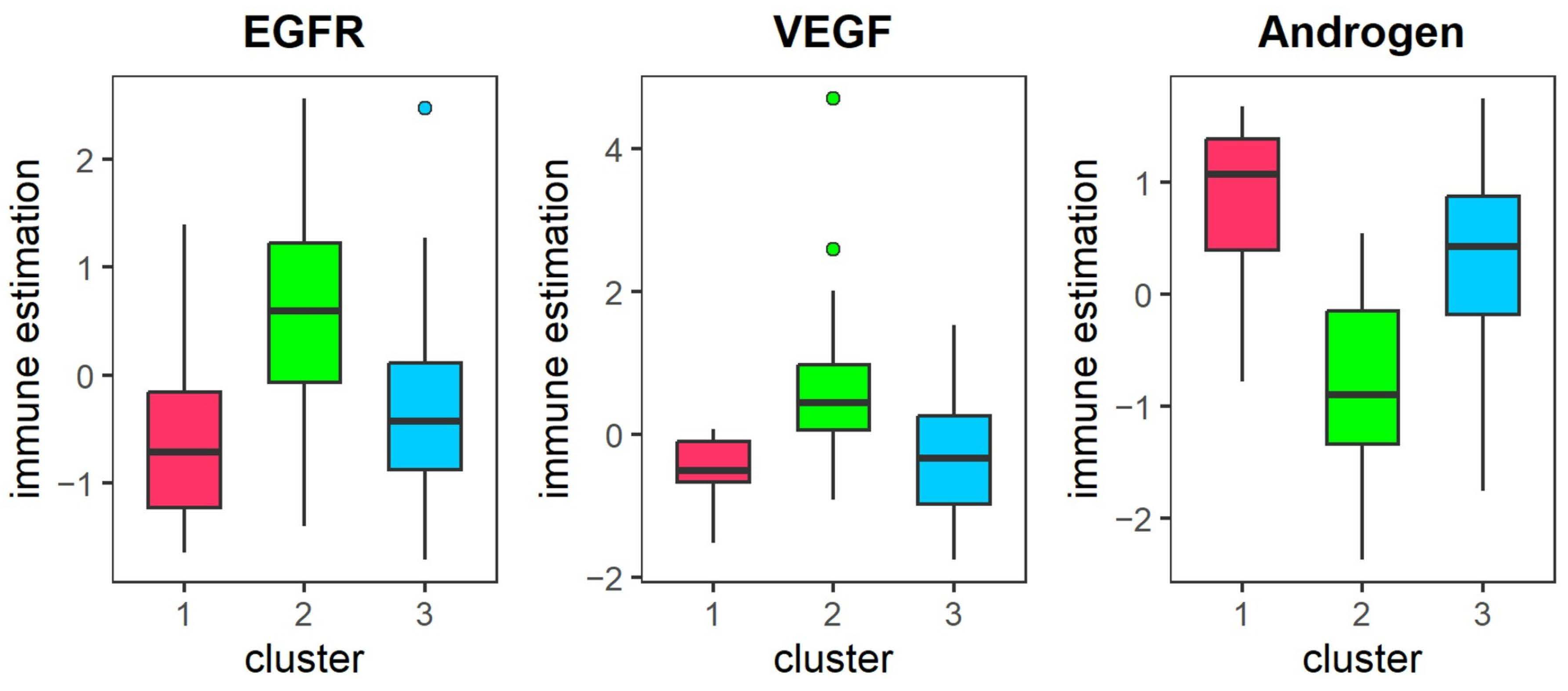
Immune cell infiltration and Pathway activity analysis: We employed the R package IOBR (version 0.99.99) [
40] to estimate tumor cell composition and identified immune infiltrating cells with significant differences between subtypes using the Kruskal–Wallis H test, with a threshold of
< 0.05. Additionally, we characterized differential pathway activities across subtypes by analyzing pathway activity scores for 14 signaling pathways based on gene expression data using the PROGENy package (version 1.30.0) [
41].