Genome-Wide Identification and Expression Analysis Under Abiotic Stress of the Lipoxygenase Gene Family in Maize (Zea mays)
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of LOX Family Genes in Maize
2.2. Chromosomal Distribution of ZmLOX Genes
2.3. Characterization of Conserved Motifs and Structural Features in ZmLOX Family
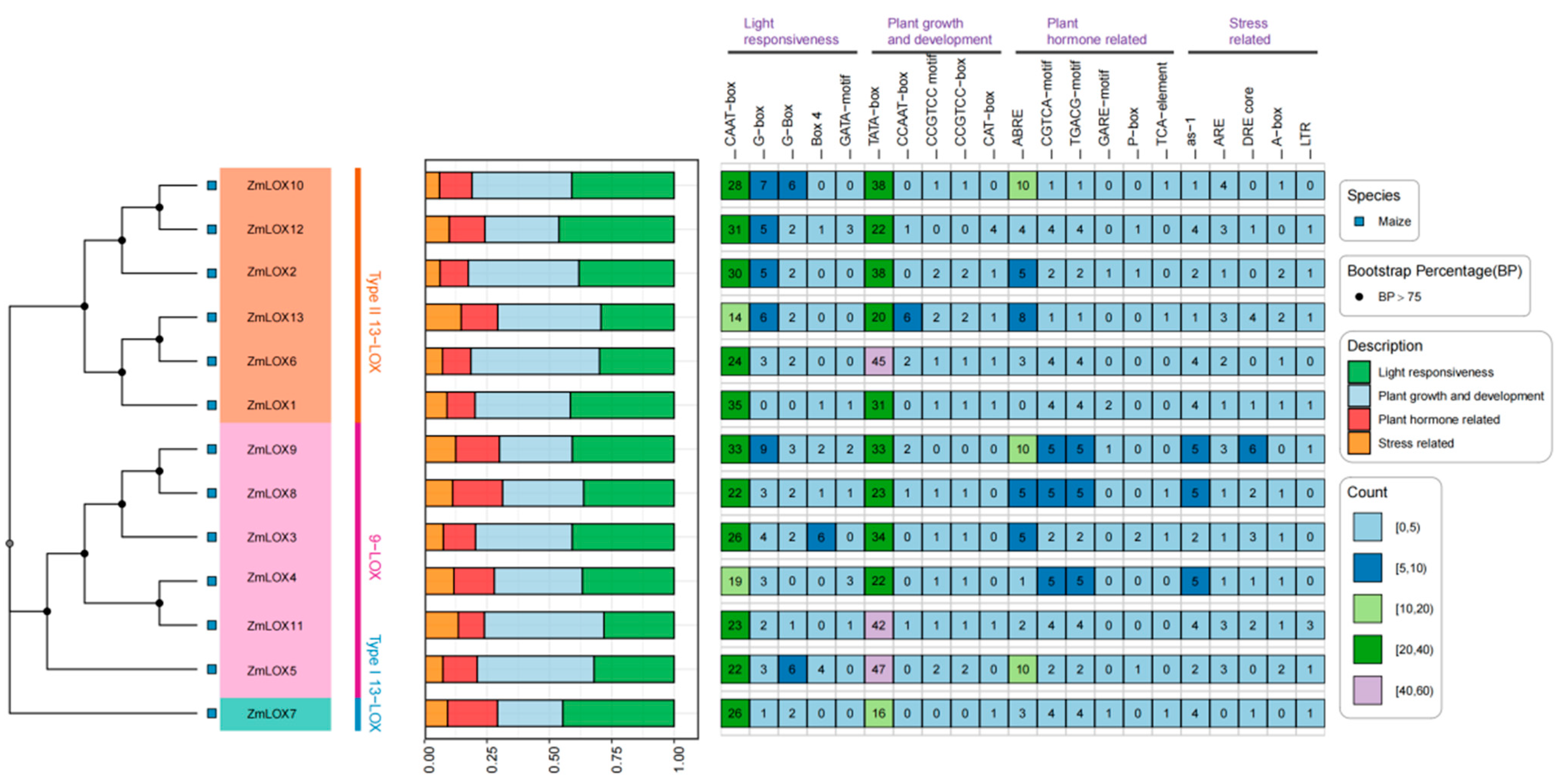
2.4. Phylogenetic Analysis of LOX Family Proteins
2.5. Collinearity Analysis of LOX Family Genes in Maize
2.6. Cis-Acting Element Analysis of ZmLOX’s Promoters
2.7. GO and KEGG Annotation of ZmLOX Genes
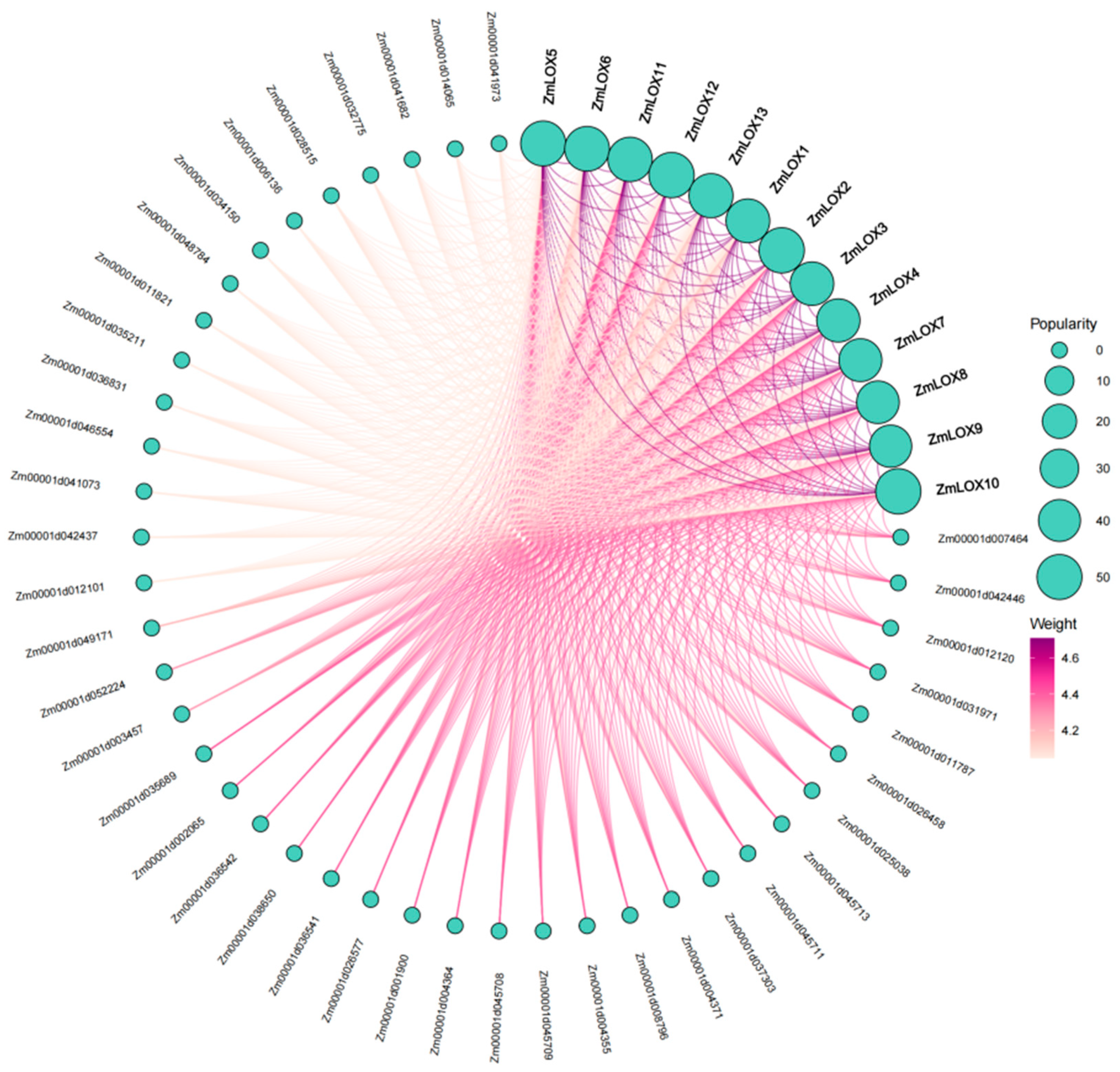
2.8. Protein–Protein Interaction Network of ZmLOX in Maize
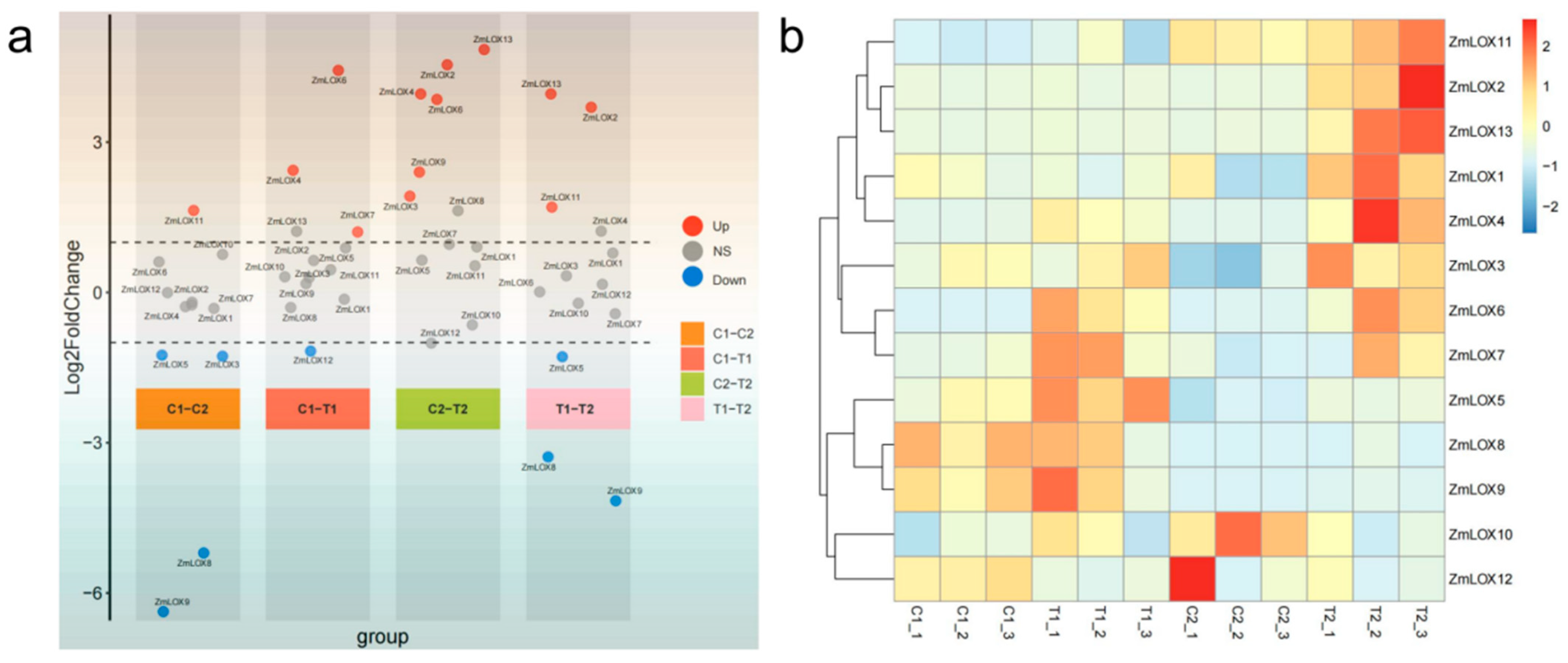
2.9. Differential Expression and Expression Profiling of ZmLOX Genes Based on Transcriptome Data
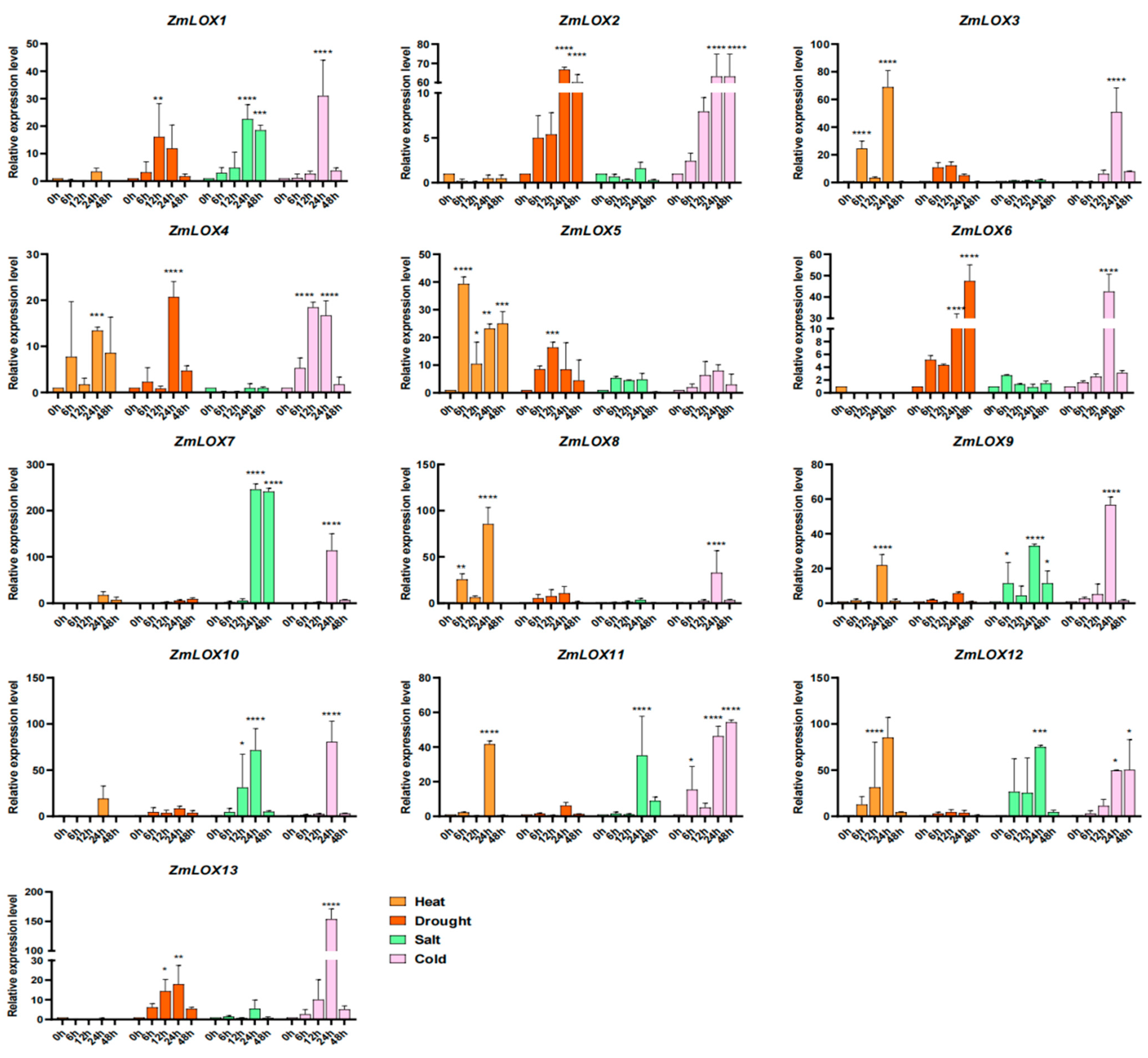
2.10. Expression Analysis of ZmLOX Family Genes Under Abiotic Stress Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Identification of LOX Family Genes in Maize
4.3. Chromosome Distribution Analysis of ZmLOX Genes
4.4. Motif and Domain Analysis of ZmLOX Proteins
4.5. Gene Structure Analysis of ZmLOX Genes
4.6. Phylogenetic Analysis of LOX Family Members
4.7. Collinearity Analysis of Maize LOX Family Genes
4.8. Cis-Acting Element Analysis of ZmLOX Gene Promoters
4.9. GO and KEGG Analysis
4.10. PPI Network Analysis
4.11. Expression Analysis of ZmLOX Genes
4.12. qRT-PCR Analysis
4.13. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Porta, H.; Rocha-Sosa, M. Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 2002, 130, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.A.; Ashraf, U.; Tanveer, M.; Khan, I.; Hussain, S.; Shahzad, B.; Zohaib, A.; Abbas, F.; Saleem, M.F.; Ali, I. Drought induced changes in growth, osmolyte accumulation and antioxidant metabolism of three maize hybrids. Front. Plant Sci. 2017, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Rohman, M.M.; Talukder, M.; Hossain, M.; Uddin, M.; Amiruzzaman, M.; Biswas, A.; Ahsan, A.; Chowdhury, M. Saline sensitivity leads to oxidative stress and increases the antioxidants in presence of proline and betaine in maize (‘Zea mays’ L.) inbred. Plant Omics 2016, 9, 35–47. [Google Scholar]
- Brash, A.R. Lipoxygenases: Occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem. 1999, 274, 23679–23682. [Google Scholar] [CrossRef] [PubMed]
- Steczko, J.; Donoho, G.P.; Clemens, J.C.; Dixon, J.E.; Axelrod, B. Conserved histidine residues in soybean lipoxygenase: Functional consequences of their replacement. Biochemistry 1992, 31, 4053–4057. [Google Scholar] [CrossRef]
- Wang, R.; Shen, W.; Liu, L.; Jiang, L.; Liu, Y.; Su, N.; Wan, J. A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Mol. Biol. 2008, 66, 401–414. [Google Scholar] [CrossRef]
- Veronico, P.; Giannino, D.; Melillo, M.T.; Leone, A.; Reyes, A.; Kennedy, M.W.; Bleve-Zacheo, T. A novel lipoxygenase in pea roots. Its function in wounding and biotic stress. Plant Physiol. 2006, 141, 1045–1055. [Google Scholar] [CrossRef][Green Version]
- Kolomiets, M.V.; Hannapel, D.J.; Chen, H.; Tymeson, M.; Gladon, R.J. Lipoxygenase is involved in the control of potato tuber development. Plant Cell 2001, 13, 613–626. [Google Scholar] [CrossRef]
- Porta, H.; Figueroa-Balderas, R.E.; Rocha-Sosa, M. Wounding and pathogen infection induce a chloroplast-targeted lipoxygenase in the common bean (Phaseolus vulgaris L.). Planta 2008, 227, 363–373. [Google Scholar] [CrossRef]
- López, M.A.; Vicente, J.; Kulasekaran, S.; Vellosillo, T.; Martínez, M.; Irigoyen, M.L.; Cascón, T.; Bannenberg, G.; Hamberg, M.; Castresana, C. Antagonistic role of 9-lipoxygenase-derived oxylipins and ethylene in the control of oxidative stress, lipid peroxidation and plant defence. Plant J. 2011, 67, 447–458. [Google Scholar] [CrossRef]
- Vellosillo, T.; Martínez, M.; Lopez, M.A.; Vicente, J.; Cascon, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, L.; Li, Z.; Wang, W.; Qi, Z.; Li, Y.; Liu, Y.; Liu, Z. Genome-Wide Identification of LOX Gene Family and Its Expression Analysis under Abiotic Stress in Potato (Solanum tuberosum L.). Int. J. Mol. Sci. 2024, 25, 3487. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Sun, Q.; Yuan, C.; Zhao, X.; Wang, J.; Yan, C.; Li, C.; Shan, S. Identification of the LOX gene family in peanut and functional characterization of AhLOX29 in drought tolerance. Front. Plant Sci. 2022, 13, 832785. [Google Scholar] [CrossRef] [PubMed]
- Shaban, M.; Ahmed, M.M.; Sun, H.; Ullah, A.; Zhu, L. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genom. 2018, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jin, Y.; Liu, J.; Tang, Y.; Cao, S.; Qi, H. The phylogeny and expression profiles of the lipoxygenase (LOX) family genes in the melon (Cucumis melo L.) genome. Sci. Hortic. 2014, 170, 94–102. [Google Scholar] [CrossRef]
- Li, M.; Li, L.; Dunwell, J.M.; Qiao, X.; Liu, X.; Zhang, S. Characterization of the lipoxygenase (LOX) gene family in the Chinese white pear (Pyrus bretschneideri) and comparison with other members of the Rosaceae. BMC Genom. 2014, 15, 444. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, K.; Bowen, J.; Allan, A.; Espley, R.; Karunairetnam, S.; Ferguson, I. Differential expression within the LOX gene family in ripening kiwifruit. J. Exp. Bot. 2006, 57, 3825–3836. [Google Scholar] [CrossRef]
- Yang, X.-Y.; Jiang, W.-J.; Yu, H.-J. The expression profiling of the lipoxygenase (LOX) family genes during fruit development, abiotic stress and hormonal treatments in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2012, 13, 2481–2500. [Google Scholar] [CrossRef]
- Huang, D.; Ma, F.; Wu, B.; Lv, W.; Xu, Y.; Xing, W.; Chen, D.; Xu, B.; Song, S. Genome-wide association and expression analysis of the lipoxygenase gene family in Passiflora edulis revealing pelox4 might be involved in fruit ripeness and ester formation. Int. J. Mol. Sci. 2022, 23, 12496. [Google Scholar] [CrossRef]
- Li, Z.; Xie, Q.; Yan, J.; Chen, J.; Chen, Q. Genome-wide identification and characterization of the abiotic-stress-responsive GRF gene family in diploid woodland strawberry (Fragaria vesca). Plants 2021, 10, 1916. [Google Scholar] [CrossRef]
- Meng, Y.; Liang, Y.; Liao, B.; He, W.; Liu, Q.; Shen, X.; Xu, J.; Chen, S. Genome-wide identification, characterization and expression analysis of lipoxygenase gene family in Artemisia annua L. Plants 2022, 11, 655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, Y.; Zhang, J.; Li, X.; Ma, F.; Duan, M.; Zhang, B.; Li, H. The responses of the lipoxygenase gene family to salt and drought stress in foxtail millet (Setaria italica). Life 2021, 11, 1169. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, L.; Chen, P.; Li, Q.; Yang, Q.; Zhang, Y.; Tan, B.; Ye, X.; Zheng, X.; Feng, J. Genome-wide identification and expression of the lipoxygenase gene family in jujube (Ziziphus jujuba) in response to phytoplasma infection. J. Plant Biochem. Biotechnol. 2022, 31, 139–153. [Google Scholar] [CrossRef]
- Shrestha, K.; Pant, S.; Huang, Y. Genome-wide identification and classification of Lipoxygenase gene family and their roles in sorghum-aphid interaction. Plant Mol. Biol. 2021, 105, 527–541. [Google Scholar] [CrossRef]
- Sarde, S.J.; Kumar, A.; Remme, R.N.; Dicke, M. Genome-wide identification, classification and expression of lipoxygenase gene family in pepper. Plant Mol. Biol. 2018, 98, 375–387. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, X.; Guo, L.; Xu, Q.; Zhao, S.; Li, F.; Yan, X.; Liu, S.; Wei, C. Characterization and alternative splicing profiles of the lipoxygenase gene family in tea plant (Camellia sinensis). Plant Cell Physiol. 2018, 59, 1765–1781. [Google Scholar] [CrossRef]
- Guo, S.; Song, Z.; Ma, R.; Yang, Y.; Yu, M. Genome-wide identification and expression analysis of the lipoxygenase gene family during peach fruit ripening under different postharvest treatments. Acta Physiol. Plant. 2017, 39, 111. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, X.; Yan, H.; Li, W.; Li, Y.; Cai, R.; Xiang, Y. The lipoxygenase gene family in poplar: Identification, classification, and expression in response to MeJA treatment. PLoS ONE 2015, 10, e0125526. [Google Scholar] [CrossRef]
- Umate, P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 2011, 6, 335–338. [Google Scholar] [CrossRef]
- Ogunola, O.F.; Hawkins, L.K.; Mylroie, E.; Kolomiets, M.V.; Borrego, E.; Tang, J.D.; Williams, W.P.; Warburton, M.L. Characterization of the maize lipoxygenase gene family in relation to aflatoxin accumulation resistance. PLoS ONE 2017, 12, e0181265. [Google Scholar] [CrossRef]
- Gong, F.; Yang, L.; Tai, F.; Hu, X.; Wang, W. “Omics” of maize stress response for sustainable food production: Opportunities and challenges. Omics A J. Integr. Biol. 2014, 18, 714–732. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Dichio, B.; Xiloyannis, C.; Masia, A. Lipoxygenase activity and proline accumulation in leaves and roots of olive trees in response to drought stress. Physiol. Plant. 2004, 121, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.-S.; Rahimi, S.; Kim, Y.-J.; Devi, B.S.R.; Khorolragchaa, A.; Sukweenadhi, J.; Silva, J.; Myagmarjav, D.; Yang, D.-C. Molecular characterization of lipoxygenase genes and their expression analysis against biotic and abiotic stresses in Panax ginseng. Eur. J. Plant Pathol. 2016, 145, 331–343. [Google Scholar] [CrossRef]
- Rai, A.N.; Mandliya, T.; Kulkarni, P.; Rao, M.; Suprasanna, P. Evolution and transcriptional modulation of lipoxygenase genes under heat, drought, and combined stress in Brassica rapa. Plant Mol. Biol. Report. 2021, 39, 60–71. [Google Scholar] [CrossRef]
- Chen, T.; Cohen, D.; Itkin, M.; Malitsky, S.; Fluhr, R. Lipoxygenase functions in 1O2 production during root responses to osmotic stress. Plant Physiol. 2021, 185, 1638–1651. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, K.; Wang, Z.; Li, X.; Li, M.; Zhu, F.; Majeed, Z.; Lan, X.; Guan, Q. The lipoxygenase gene AfLOX4 of Amorpha fruticosa L. is a potential regulator of drought stress tolerance pathways under saline and alkaline conditions. Acta Physiol. Plant. 2023, 45, 72. [Google Scholar] [CrossRef]
- Lian, C.; Li, Q.; Yao, K.; Zhang, Y.; Meng, S.; Yin, W.; Xia, X. Populus trichocarpa PtNF-YA9, a multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis. Front. Plant Sci. 2018, 9, 954. [Google Scholar] [CrossRef]
- Lv, R.; Mo, F.; Li, C.; Meng, F.; Zhang, H.; Yu, L.; Cheng, M.; Wang, P.; Liu, S.; Liu, Z. Genome-wide identification of the CLC gene family in tomato (Solanum lycopersicum) and functional analysis of SlCLC8 in salt stress tolerance. Sci. Hortic. 2024, 338, 113754. [Google Scholar] [CrossRef]
- Mo, F.; Xue, X.; Meng, L.; Zhang, Y.; Cui, Y.; Liu, J.; Cheng, M.; Wang, P.; Lv, R.; Meng, F. Genome-wide identification and expression analysis of SLAC1 gene family in tomato (Solanum lycopersicum) and the function of SlSLAC1–6 under cold stress. Sci. Hortic. 2023, 313, 111904. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Krishnakumar, V.; Zeng, X.; Xu, Z.; Taranto, A.; Lomas, J.S.; Zhang, Y.; Huang, Y.; Wang, Y.; Yim, W.C. JCVI: A versatile toolkit for comparative genomics analysis. iMeta 2024, 3, e211. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
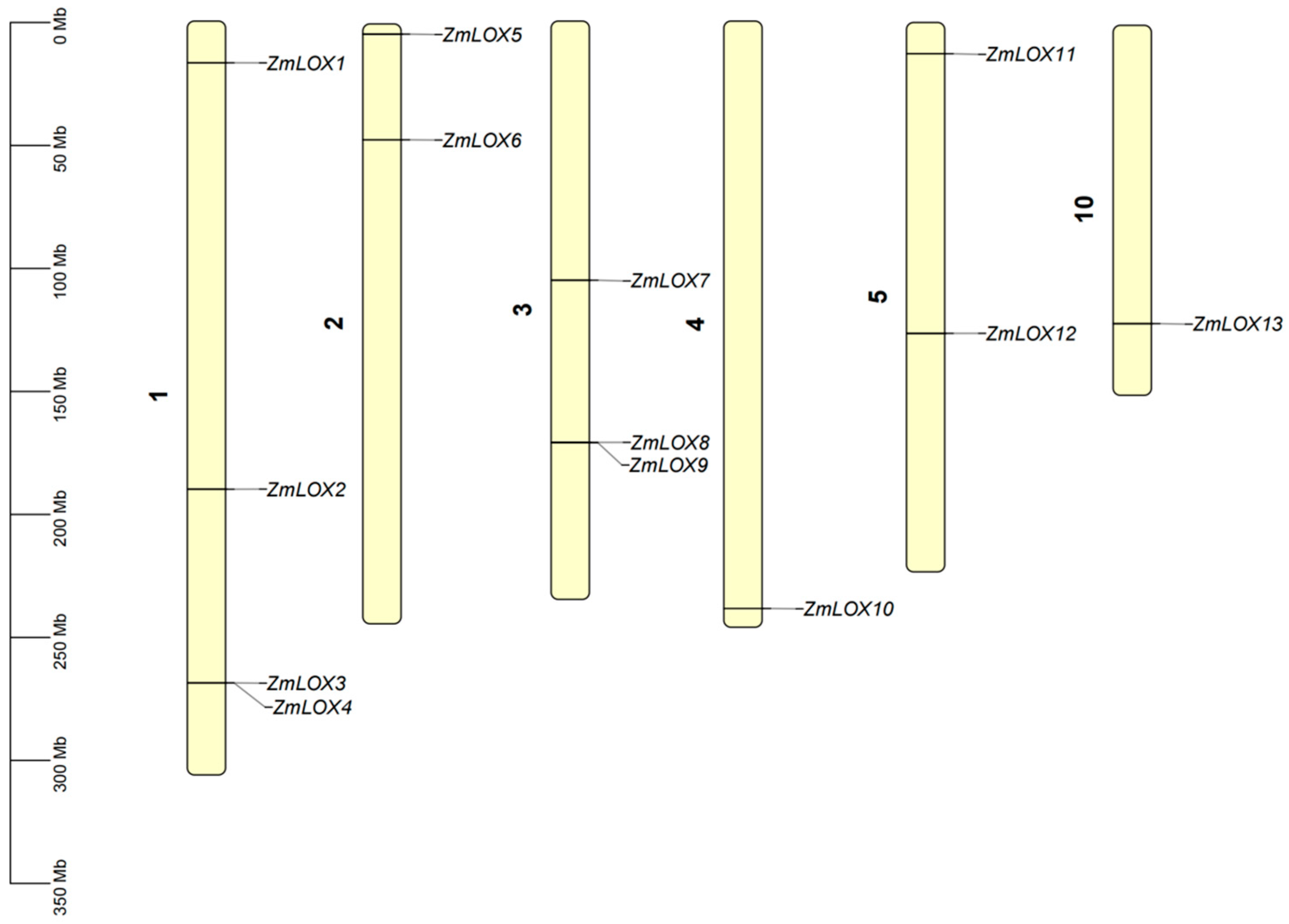
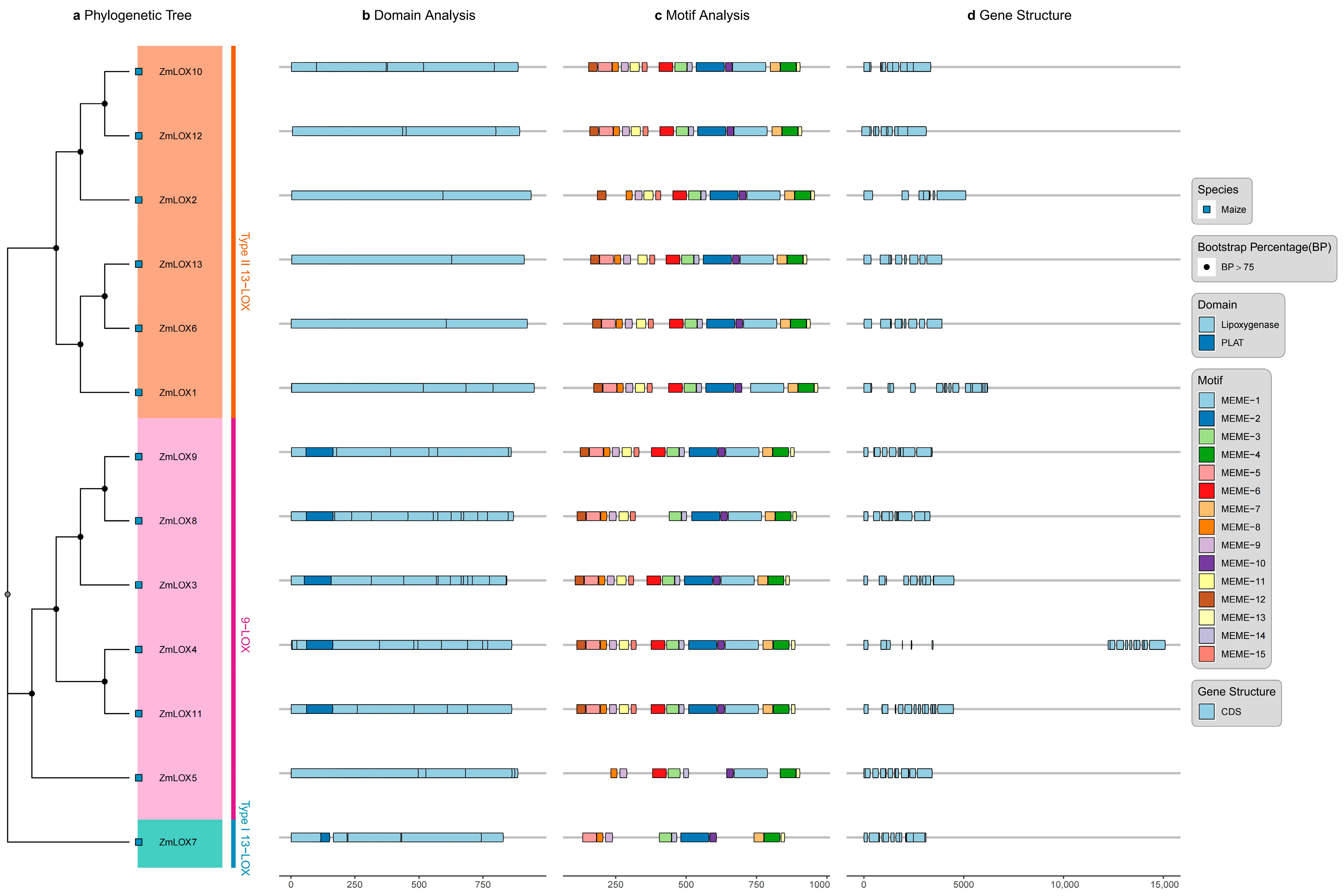
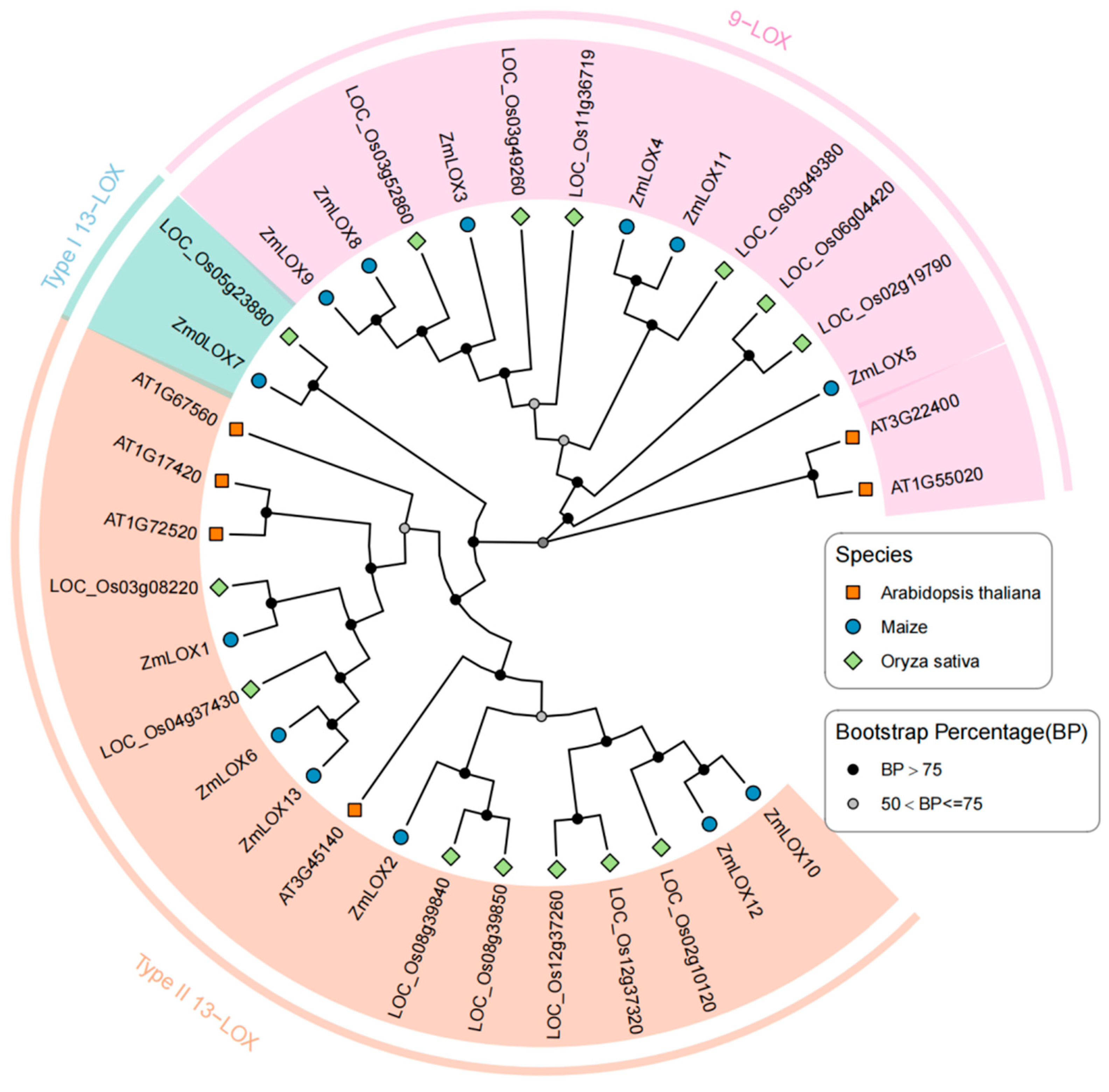
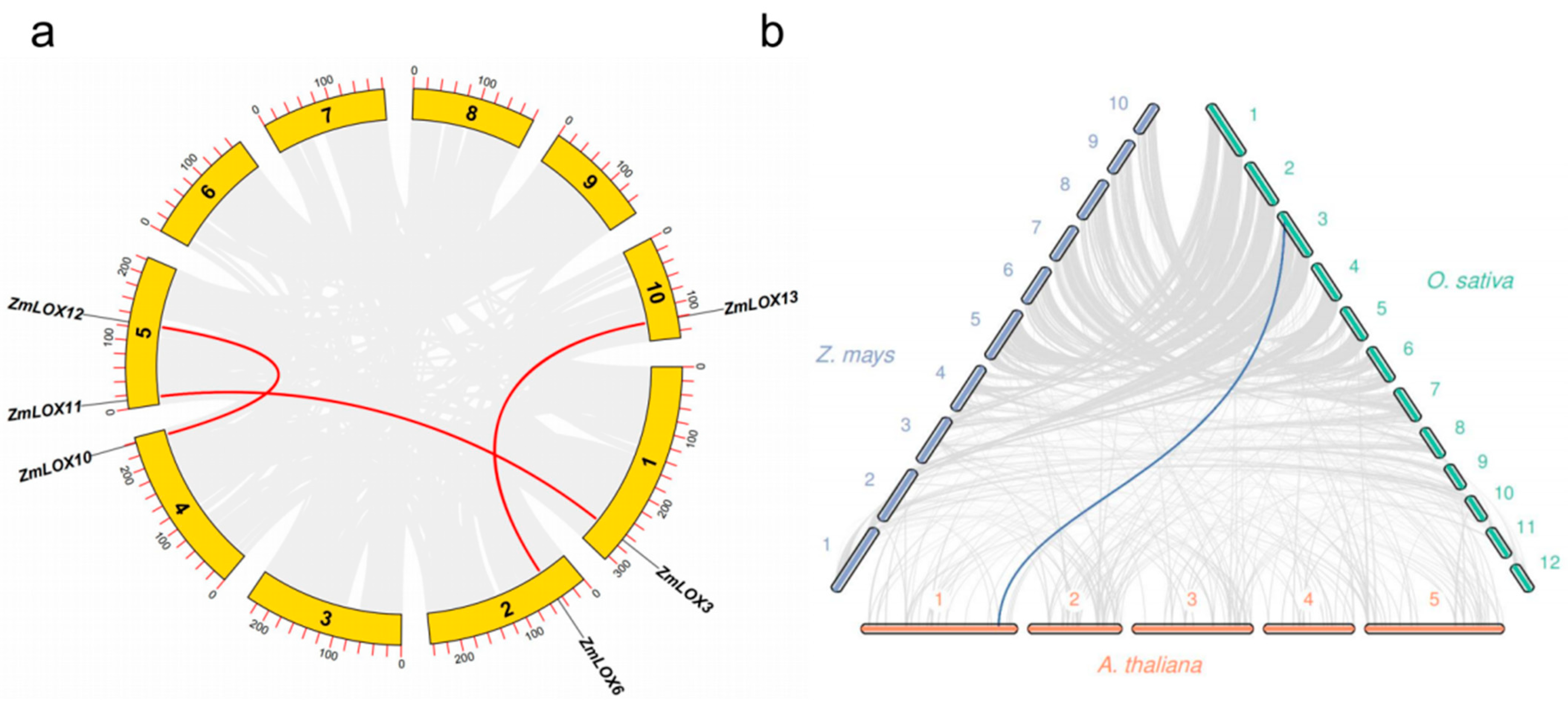

| Name | ID | Chr | Start | End | MW (KDa) | Length | Hydrophobicity | pI | Cell |
|---|---|---|---|---|---|---|---|---|---|
| ZmLOX1 | Zm00001d027893 | 1 | 16948608 | 16955122 | 108.27 | 968 | −0.33 | 8.59 | Cytoplasm |
| ZmLOX2 | Zm00001d031449 | 1 | 190316537 | 190321767 | 105.33 | 956 | −0.33 | 6.73 | Chloroplast |
| ZmLOX3 | Zm00001d033623 | 1 | 269047817 | 269052874 | 96.73 | 867 | −0.32 | 6.06 | Cytoplasm |
| ZmLOX4 | Zm00001d033624 | 1 | 269056560 | 269072120 | 100.36 | 887 | −0.45 | 6.63 | Cytoplasm |
| ZmLOX5 | Zm00001d002000 | 2 | 4150293 | 4154404 | 98.66 | 904 | −0.29 | 6.00 | Cytoplasm |
| ZmLOX6 | Zm00001d003533 | 2 | 47105187 | 47109372 | 105.31 | 941 | −0.44 | 7.28 | Cytoplasm |
| ZmLOX7 | Zm00001d041204 | 3 | 105356377 | 105359732 | 95.99 | 850 | −0.44 | 8.22 | Cytoplasm |
| ZmLOX8 | Zm00001d042540 | 3 | 171277704 | 171281453 | 100.75 | 892 | −0.37 | 6.68 | Cytoplasm |
| ZmLOX9 | Zm00001d042541 | 3 | 171421717 | 171425401 | 99.33 | 884 | −0.34 | 6.81 | Cytoplasm |
| ZmLOX10 | Zm00001d053675 | 4 | 238805319 | 238809230 | 102.06 | 905 | −0.39 | 6.54 | Chloroplast |
| ZmLOX11 | Zm00001d013493 | 5 | 12701180 | 12706156 | 100.36 | 887 | −0.43 | 6.83 | Cytoplasm |
| ZmLOX12 | Zm00001d015852 | 5 | 126372618 | 126376084 | 102.87 | 911 | −0.39 | 7.04 | Chloroplast |
| ZmLOX13 | Zm00001d025524 | 10 | 121268606 | 121272616 | 103.6 | 929 | −0.42 | 7.14 | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Hou, S.; Sun, Y.; Sun, M.; Sun, Y.; Li, X.; Li, Y.; Wang, L.; Cai, Q.; Guo, B.; et al. Genome-Wide Identification and Expression Analysis Under Abiotic Stress of the Lipoxygenase Gene Family in Maize (Zea mays). Genes 2025, 16, 99. https://doi.org/10.3390/genes16010099
Li S, Hou S, Sun Y, Sun M, Sun Y, Li X, Li Y, Wang L, Cai Q, Guo B, et al. Genome-Wide Identification and Expression Analysis Under Abiotic Stress of the Lipoxygenase Gene Family in Maize (Zea mays). Genes. 2025; 16(1):99. https://doi.org/10.3390/genes16010099
Chicago/Turabian StyleLi, Sinan, Shuai Hou, Yuanqing Sun, Minghao Sun, Yan Sun, Xin Li, Yunlong Li, Luyao Wang, Quan Cai, Baitao Guo, and et al. 2025. "Genome-Wide Identification and Expression Analysis Under Abiotic Stress of the Lipoxygenase Gene Family in Maize (Zea mays)" Genes 16, no. 1: 99. https://doi.org/10.3390/genes16010099
APA StyleLi, S., Hou, S., Sun, Y., Sun, M., Sun, Y., Li, X., Li, Y., Wang, L., Cai, Q., Guo, B., & Zhang, J. (2025). Genome-Wide Identification and Expression Analysis Under Abiotic Stress of the Lipoxygenase Gene Family in Maize (Zea mays). Genes, 16(1), 99. https://doi.org/10.3390/genes16010099






