The Prevalence and Clinical Characteristics of MYO3A-Associated Hearing Loss in 15,684 Hearing Loss Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Variant Analysis
2.3. Clinical Evaluations
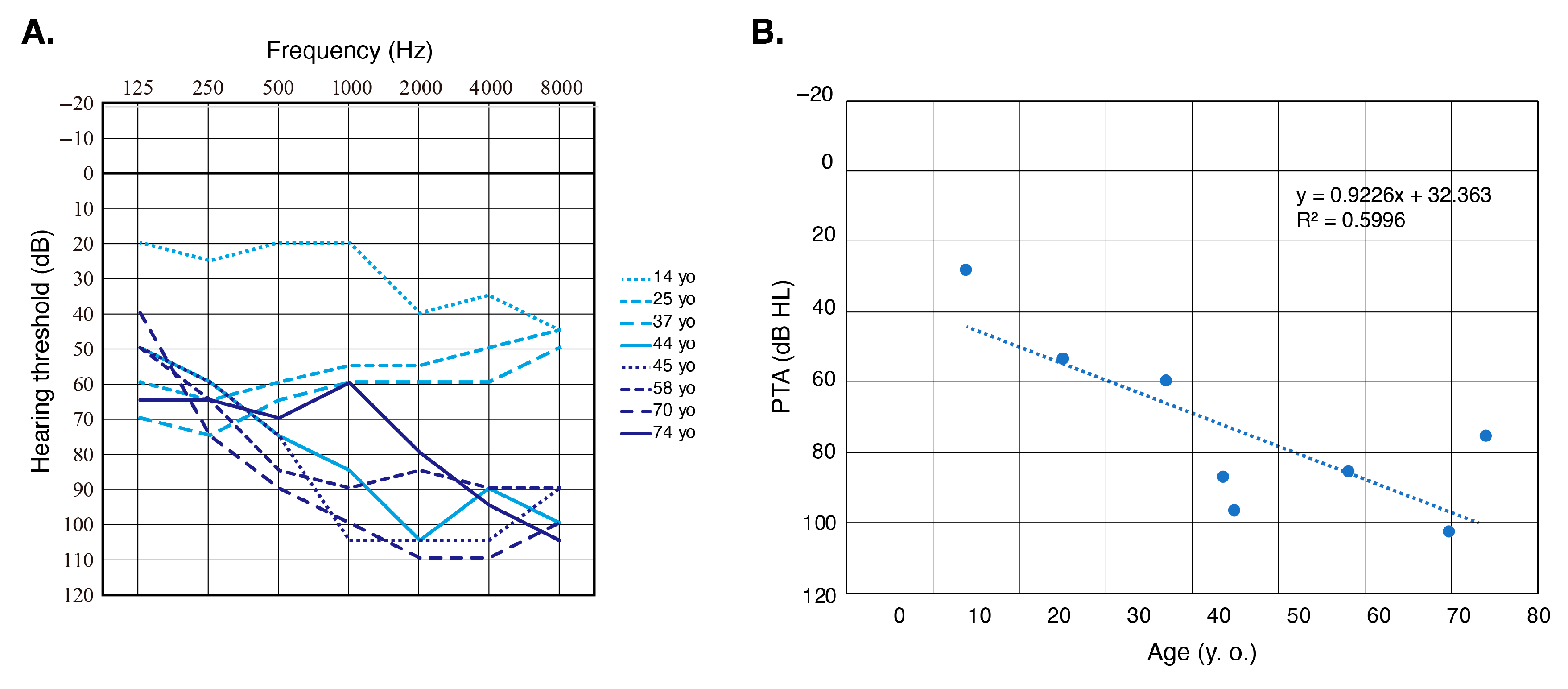
3. Results
3.1. Detected Variations
3.2. Clinical Characteristics of MYO3A-Associated HL
4. Discussion
| Nucleotide Change | AA Change | Inheritance | Onset | Severity | Configuration | Progression | Population | Reference |
|---|---|---|---|---|---|---|---|---|
| c.[149A>G];[149A>G] | p.[K50R];[K50R] | AR | Congenital | Profound | NA | NA | Tunisia | [26] |
| c.[424C>T];[424C>T] | p.[H142Y];[H142Y] | AR | Congenital/ Pre-lingual | Severe-to-profound | NA | NA | South Africa | [23] |
| c.[580C>A];[1582_1583insT] | p.[P194T];[Y530Lfs*9] | AR | NA | Severe-to-profound | HF_precipitous | NA | Korea | [18] |
| c.[732-2A>G];[732-2A>G] | p.[IVS8 as A-G -2];[IVS8 as A-G -2] | AR | Second decade | Moderate-to-profound | HF_precipitous | NA | US | [17] |
| c.[824G>A];[3737_3738delAG] | p.[R275H];[E1246Gfs*5] | AR | NA | Moderate-to-severe | NA | NA | China | [24] |
| c.[1370_1371delGA];[1370_1371delGA] | p.[R457Nfs*25];[R457Nfs*25] | AR | Pre-lingual | NA | NA | Yes | Palestine | [22] |
| c.[1450T>C];[3093G>A] | p.[S484P];[W1031X] | AR | 6 yo | Severe | HF_gentle | Yes | China | [25] |
| c.[1777-12G>A];[3126T>G] | p.[IVS17 as G-A -12];[Y1042X] | AR | Second decade | Moderate-to-profound | HF_precipitous | NA | US | [17] |
| c.[1841C>T];[1841C>T} | p.[S614F];[S614F] | AR | Congenital | Profound | NA | No | China | [20] |
| c.[3126T>G];[3126T>G] | p.[Y1042X];[Y1042X] | AR | Second decade | Moderate-to-profound | HF_precipitous | NA | US | [17] |
| c.[4462A>G];[[4681C>T] | p.[K1488E];[R1561X] | AR | Congenital/ Pre-lingual | Profound | NA | NA | Taiwan | [19] |
| c.716T>C | p.L239P | AD | Pre-lingual | Moderate-to-profound | HF_gentle | Yes | Europe | [38] |
| c.1463G>A | p.G488E | AD | Post-lingual early-onset | Moderate-to-profound | NA | Yes | African American | [36] |
| c.2090T>G | p.L697W | AD | 10-60 | Mild-to-profound | HF_gentle | Yes | Brazil. Portugal | [37] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoshimura, H.; Okubo, T.; Shinagawa, J.; Nishio, S.Y.; Takumi, Y.; Usami, S.I. Epidemiology, aetiology and diagnosis of congenital hearing loss via hearing screening of 153913 newborns. Int. J. Epidemiol. 2024, 53, dyae052. [Google Scholar] [CrossRef] [PubMed]
- Sheffield, A.M.; Smith, R.J.H. The Epidemiology of Deafness. Cold Spring Harb. Perspect. Med. 2019, 9, a033258. [Google Scholar] [CrossRef]
- Hereditary Hearing Loss Homepage. Available online: https://hereditaryhearingloss.org (accessed on 20 November 2024).
- Aldè, M.; Cantarella, G.; Zanetti, D.; Pignataro, L.; La Mantia, I.; Maiolino, L.; Ferlito, S.; Di Mauro, P.; Cocuzza, S.; Lechien, J.R.; et al. Autosomal Dominant Non-Syndromic Hearing Loss (DFNA): A Comprehensive Narrative Review. Biomedicines 2023, 11, 1616. [Google Scholar] [CrossRef] [PubMed]
- Usami, S.I.; Nishio, S.Y. The genetic etiology of hearing loss in Japan revealed by the social health insurance-based genetic testing of 10K patients. Hum. Genet. 2022, 141, 665–681. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Quintero, O.A.; Moore, J.E.; Unrath, W.C.; Manor, U.; Salles, F.T.; Grati, M.; Kachar, B.; Yengo, C.M. Intermolecular autophosphorylation regulates myosin IIIa activity and localization in parallel actin bundles. J. Biol. Chem. 2010, 285, 35770–35782. [Google Scholar] [CrossRef] [PubMed]
- Quintero, O.A.; Unrath, W.C.; Stevens, S.M., Jr.; Manor, U.; Kachar, B.; Yengo, C.M. Myosin 3A kinase activity is regulated by phosphorylation of the kinase domain activation loop. J. Biol. Chem. 2013, 288, 37126–37137. [Google Scholar] [CrossRef] [PubMed]
- Salles, F.T.; Merritt, R.C., Jr.; Manor, U.; Dougherty, G.W.; Sousa, A.D.; Moore, J.E.; Yengo, C.M.; Dosé, A.C.; Kachar, B. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat. Cell Biol. 2009, 11, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Lelli, A.; Michel, V.; Boutet de Monvel, J.; Cortese, M.; Bosch-Grau, M.; Aghaie, A.; Perfettini, I.; Dupont, T.; Avan, P.; El-Amraoui, A.; et al. Class III myosins shape the auditory hair bundles by limiting microvilli and stereocilia growth. J. Cell Biol. 2016, 212, 231–244. [Google Scholar] [CrossRef]
- Merritt, R.C.; Manor, U.; Salles, F.T.; Grati, M.; Dose, A.C.; Unrath, W.C.; Quintero, O.A.; Yengo, C.M.; Kachar, B. Myosin IIIB uses an actin-binding motif in its espin-1 cargo to reach the tips of actin protrusions. Curr. Biol. 2012, 22, 320–325. [Google Scholar] [CrossRef]
- Dosé, A.C.; Hillman, D.W.; Wong, C.; Sohlberg, L.; Lin-Jones, J.; Burnside, B. Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus. Mol Biol. Cell 2003, 14, 1058–1073. [Google Scholar] [CrossRef] [PubMed]
- HGMD Professional. Available online: http://www.hgmd.org (accessed on 20 November 2024).
- Cirilo, J.A., Jr.; Gunther, L.K.; Yengo, C.M. Functional Role of Class III Myosins in Hair Cells. Front. Cell Dev. Biol. 2021, 25, 643856. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, S.; Avenarius, M.R.; Grati, M.; Krey, J.F.; Windsor, A.M.; Sousa, A.D.; Ballesteros, A.; Cui, R.; Millis, B.A.; Salles, F.T.; et al. Stereocilia-staircase spacing is influenced by myosin III motors and their cargos espin-1 and espin-like. Nat. Commun. 2016, 1, 10833. [Google Scholar] [CrossRef] [PubMed]
- Les Erickson, F.; Corsa, A.C.; Dosé, A.C.; Burnside, B. Localization of a class III myosin to filopodia tips in transfected HeLa cells requires an actin-binding site in its tail domain. Mol. Biol. Cell 2003, 14, 4173–4180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Walsh, T.; Walsh, V.; Vreugde, S.; Hertzano, R.; Shahin, H.; Haika, S.; Lee, M.K.; Kanaan, M.; King, M.C.; Avraham, K.B. From flies’ eyes to our ears: Mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Natl. Acad. Sci. USA 2002, 99, 7518–7523. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.Y.; Park, G.; Gim, J.; Kim, A.R.; Kim, B.J.; Kim, H.S.; Park, J.H.; Park, T.; Oh, S.H.; Han, K.H.; et al. Diagnostic application of targeted resequencing for familial nonsyndromic hearing loss. PLoS ONE 2013, 8, e68692. [Google Scholar] [CrossRef]
- Wu, C.C.; Lin, Y.H.; Liu, T.C.; Lin, K.N.; Yang, W.S.; Hsu, C.J.; Chen, P.L.; Wu, C.M. Identifying children with poor cochlear implantation outcomes using massively parallel sequencing. Medicine 2015, 94, e1073. [Google Scholar] [CrossRef]
- Sommen, M.; Schrauwen, I.; Vandeweyer, G.; Boeckx, N.; Corneveaux, J.J.; van den Ende, J.; Boudewyns, A.; De Leenheer, E.; Janssens, S.; Claes, K.; et al. DNA Diagnostics of Hereditary Hearing Loss: A Targeted Resequencing Approach Combined with a Mutation Classification System. Hum. Mutat. 2016, 37, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Qu, R.; Sang, Q.; Xu, Y.; Feng, R.; Jin, L.; He, L.; Wang, L. Identification of a novel homozygous mutation in MYO3A in a Chinese family with DFNB30 non-syndromic hearing impairment. Int. J. Pediatr. Otorhinolaryngol. 2016, 84, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Abu Rayyan, A.; Kamal, L.; Casadei, S.; Brownstein, Z.; Zahdeh, F.; Shahin, H.; Canavati, C.; Dweik, D.; Jaraysa, T.; Rabie, G.; et al. Genomic analysis of inherited hearing loss in the Palestinian population. Proc. Natl. Acad. Sci. USA 2020, 117, 20070–20076. [Google Scholar] [CrossRef]
- Wonkam, A.; Manyisa, N.; Bope, C.D.; Dandara, C.; Chimusa, E.R. Whole exome sequencing reveals pathogenic variants in MYO3A, MYO15A and COL9A3 and differential frequencies in ancestral alleles in hearing impairment genes among individuals from Cameroon. Hum. Mol. Genet. 2021, 29, 3729–3743. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cao, Z.; Su, Y.; Wang, Y.; Cai, R.; Chen, J.; Gao, B.; Han, M.; Li, X.; Zhang, D.; et al. Molecular diagnose of a large hearing loss population from China by targeted genome sequencing. J. Hum. Genet. 2022, 67, 643–649. [Google Scholar] [CrossRef]
- Guan, J.; Li, J.; Chen, G.; Shi, T.; Lan, L.; Wu, X.; Zhao, C.; Wang, D.; Wang, H.; Wang, Q. Family trio-based sequencing in 404 sporadic bilateral hearing loss patients discovers recessive and De novo genetic variants in multiple ways. Eur. J. Med. Genet. 2021, 64, 104311. [Google Scholar] [CrossRef] [PubMed]
- Souissi, A.; Abdelmalek Driss, D.; Chakchouk, I.; Ben Said, M.; Ben Ayed, I.; Mosrati, M.A.; Elloumi, I.; Tlili, A.; Aifa, S.; Masmoudi, S. Molecular insights into MYO3A kinase domain variants explain variability in both severity and progression of DFNB30 hearing impairment. J. Biomol. Struct. Dyn. 2022, 40, 10940–10951. [Google Scholar] [CrossRef] [PubMed]
- Nishio, S.Y.; Hayashi, Y.; Watanabe, M.; Usami, S.I. Clinical application of a custom AmpliSeq library and ion torrent PGM sequencing to comprehensive mutation screening for deafness genes. Genet. Test Mol. Biomark. 2015, 19, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids. Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- 1000 Genome Project. Available online: https://www.internationalgenome.org/1000-genomes-summary/ (accessed on 10 April 2020).
- Genome Aggregation Database. Available online: https://gnomad.broadinstitute.org (accessed on 11 November 2024).
- ToMMo 54KJPN-Integrative Japanese Genome Variation Database. Available online: https://jmorp.megabank.tohoku.ac.jp (accessed on 9 July 2023).
- Nishio, S.Y.; Moteki, H.; Usami, S.I. Simple and efficient germline copy number variant visualization method for the Ion AmpliSeq custom panel. Mol. Genet. Genom. Med. 2018, 6, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Oza, A.M.; DiStefano, M.T.; Hemphill, S.E.; Cushman, B.J.; Grant, A.R.; Siegert, R.K.; Shen, J.; Chapin, A.; Boczek, N.J.; Schimmenti, L.A.; et al. ClinGen Hearing Loss Clinical Domain Working Group. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018, 39, 1593–1613. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, M.; Van Camp, G.; Newton, V.; Giarbini, N.; Declau, F.; Parving, A. Recommendations for the Description of Genetic and Audiological Data for Families with Nonsyndromic Hereditary Hearing Impairment. Audiol. Med. 2003, 1, 148–150. [Google Scholar]
- Grati, M.; Yan, D.; Raval, M.H.; Walsh, T.; Ma, Q.; Chakchouk, I.; Kannan-Sundhari, A.; Mittal, R.; Masmoudi, S.; Blanton, S.H.; et al. MYO3A causes human dominant deafness and interacts with Protocadherin 15-CD2 isoform. Hum. Mutat. 2016, 37, 481–487. [Google Scholar] [CrossRef]
- Dantas, V.G.L.; Raval, M.H.; Ballesteros, A.; Cui, R.; Gunther, L.K.; Yamamoto, G.L.; Alves, L.U.; Bueno, A.S.; Lezirovitz, K.; Pirana, S.; et al. Characterization of a novel MYO3A missense mutation associated with a dominant form of late onset hearing loss. Sci. Rep. 2018, 8, 8706. [Google Scholar] [CrossRef] [PubMed]
- Doll, J.; Hofrichter, M.A.H.; Bahena, P.; Heihoff, A.; Segebarth, D.; Müller, T.; Dittrich, M.; Haaf, T.; Vona, B. A novel missense variant in MYO3A is associated with autosomal dominant high-frequency hearing loss in a German family. Mol. Genet. Genomic. Med. 2020, 8, e1343. [Google Scholar] [CrossRef] [PubMed]


| Nucleotide Change | AA Change | Exon | Domain | SIFT | PP2 | LRT | MutTaster | MutAssessor | REVEL | CADD | ToMMo 54KJPN | gnomAD All | Pathogenicity | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.409-2A>- | exon6 | Kinase | . | . | . | . | . | . | . | 1.80E-05 | . | Likely pathogenic (PVS1, PM2) | This study | |
| c.712G>A | p.Ala238Thr | exon8 | Kinase | T | D | D | D | L | 0.451 | 33 | . | . | VUS (PM2, PM3_Supporting) | This study |
| c.893dup | p.Gln300Thrfs*21 | exon10 | Motor | . | . | . | . | . | . | . | 0.000221 | . | Pathogenic (PVS1, PM2_Supporting, PM3_Supporting) | This study |
| c.991C>T | p.Arg331* | exon11 | Motor | . | . | D | A | . | . | . | 9.00 × 10−6 | 3.18 × 10−5 | Pathogenic (PVS1, PM2_Supporting, PM3_Supporting) | [20] |
| c.1450T>C | p.Ser484Pro | exon15 | Motor | D | D | D | D | H | 0.905 | 27.8 | 4.60 × 10−5 | 3.98 × 10−6 | Pathogenic (PM3_Strong, PP1_Strong, PM2_Supporting, PP3) | [25] |
| c.1464del | p.Lys489Asnfs*3 | exon15 | Motor | . | . | . | . | . | . | . | 0.000129 | . | Likely pathogenic (PVS1, PM2_Supporting) | This study |
| c.2308G>A | p.Glu770Lys | exon21 | Motor | T | P | D | D | L | 0.491 | 24 | 1.80 × 10−5 | . | VUS (PM2) | This study |
| c.4164dup | p.Asn1389Lysfs*4 | exon30 | Tail | . | . | . | . | . | . | . | 0.000101 | . | Likely pathogenic (PVS1, PM2_Supporting) | This study |
| Family Number | ID | Base Change Allele 1 | AA Change Allele 1 | Base Change Allele 2 | AA Change Allele 2 | Inheritance Pattern | Onset | Age | Gender | Severity of HL | Type of HL | Progression | Vestibular Symptoms |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | JHLB-3058 | c.409-2A>- | c.2308G>A | p.Glu770Lys | Sporadic | 12 | 15 | M | Moderate | HF_gentle | Y | Y | |
| 2 | JHLB-14312 | c.893dupA | p.Gln300Thrfs*21 | c.893dupA | p.Gln300Thrfs*21 | Sporadic | 22 | 74 | F | Severe | HF_gentle | Y | N |
| 3 | 2402 | c.991C>T | p.Arg331* | c.991C>T | p.Arg331* | Unknown | NA | NA | F | NA | NA | NA | NA |
| 4 | O-3349 | c.712G>A | p.Ala238Thr | c.712G>A | p.Ala238Thr | AD/Mit | 13 | 37 | F | Moderate | Flat | Y | Y |
| 5 | JHLB-5865 | c.1450T>C | p.Ser484Pro | c.1450T>C | p.Ser484Pro | AR | 20 | 58 | F | Profound | HF_gentle | Y | N |
| 6 | JHLB-12633 | c.1450T>C | p.Ser484Pro | c.1450T>C | p.Ser484Pro | AD/Mit | 25 | 44 | F | Profound | HF_gentle | Y | N |
| 7 | JHLB-13481 | c.1450T>C | p.Ser484Pro | c.1450T>C | p.Ser484Pro | Unknown | 25 | 45 | F | Profound | HF_gentle | Y | N |
| 8 | JHLB-14690 | c.1450T>C | p.Ser484Pro | c.1450T>C | p.Ser484Pro | Sporadic | 30 | 70 | F | Profound | HF_gentle | Y | N |
| 9 | JHLB-889 | c.1464delA | p.Lys489Asnfs*3 | c.4164dupA | p.Asn1389Lysfs*4 | Sporadic | 10 | 25 | F | Moderate | Flat | Y | N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maekawa, K.; Nishio, S.-y.; Miyazaki, H.; Ohta, Y.; Oishi, N.; Kasai, M.; Yamamoto, A.; Okami, M.; Wasano, K.; Sakai, A.; et al. The Prevalence and Clinical Characteristics of MYO3A-Associated Hearing Loss in 15,684 Hearing Loss Patients. Genes 2025, 16, 92. https://doi.org/10.3390/genes16010092
Maekawa K, Nishio S-y, Miyazaki H, Ohta Y, Oishi N, Kasai M, Yamamoto A, Okami M, Wasano K, Sakai A, et al. The Prevalence and Clinical Characteristics of MYO3A-Associated Hearing Loss in 15,684 Hearing Loss Patients. Genes. 2025; 16(1):92. https://doi.org/10.3390/genes16010092
Chicago/Turabian StyleMaekawa, Karuna, Shin-ya Nishio, Hiromitsu Miyazaki, Yoko Ohta, Naoki Oishi, Misato Kasai, Ai Yamamoto, Mayuri Okami, Koichiro Wasano, Akihiro Sakai, and et al. 2025. "The Prevalence and Clinical Characteristics of MYO3A-Associated Hearing Loss in 15,684 Hearing Loss Patients" Genes 16, no. 1: 92. https://doi.org/10.3390/genes16010092
APA StyleMaekawa, K., Nishio, S.-y., Miyazaki, H., Ohta, Y., Oishi, N., Kasai, M., Yamamoto, A., Okami, M., Wasano, K., Sakai, A., & Usami, S.-i. (2025). The Prevalence and Clinical Characteristics of MYO3A-Associated Hearing Loss in 15,684 Hearing Loss Patients. Genes, 16(1), 92. https://doi.org/10.3390/genes16010092







