Evolutionary and Structural Insights into the RNA Polymerase I A34 Protein Family: A Focus on Intrinsic Disorder and Phase Separation
Abstract
1. Introduction
2. Materials and Methods
2.1. Prediction of Intrinsic Disorder
2.2. Protein Structure and Function Prediction
2.3. Evolutionary Conservation Analysis
2.4. Sequence Alignment and Comparison
2.5. Sequence Logo and Motif Visualization
2.6. Phase Separation and Aggregation Propensity
3. Results and Discussion
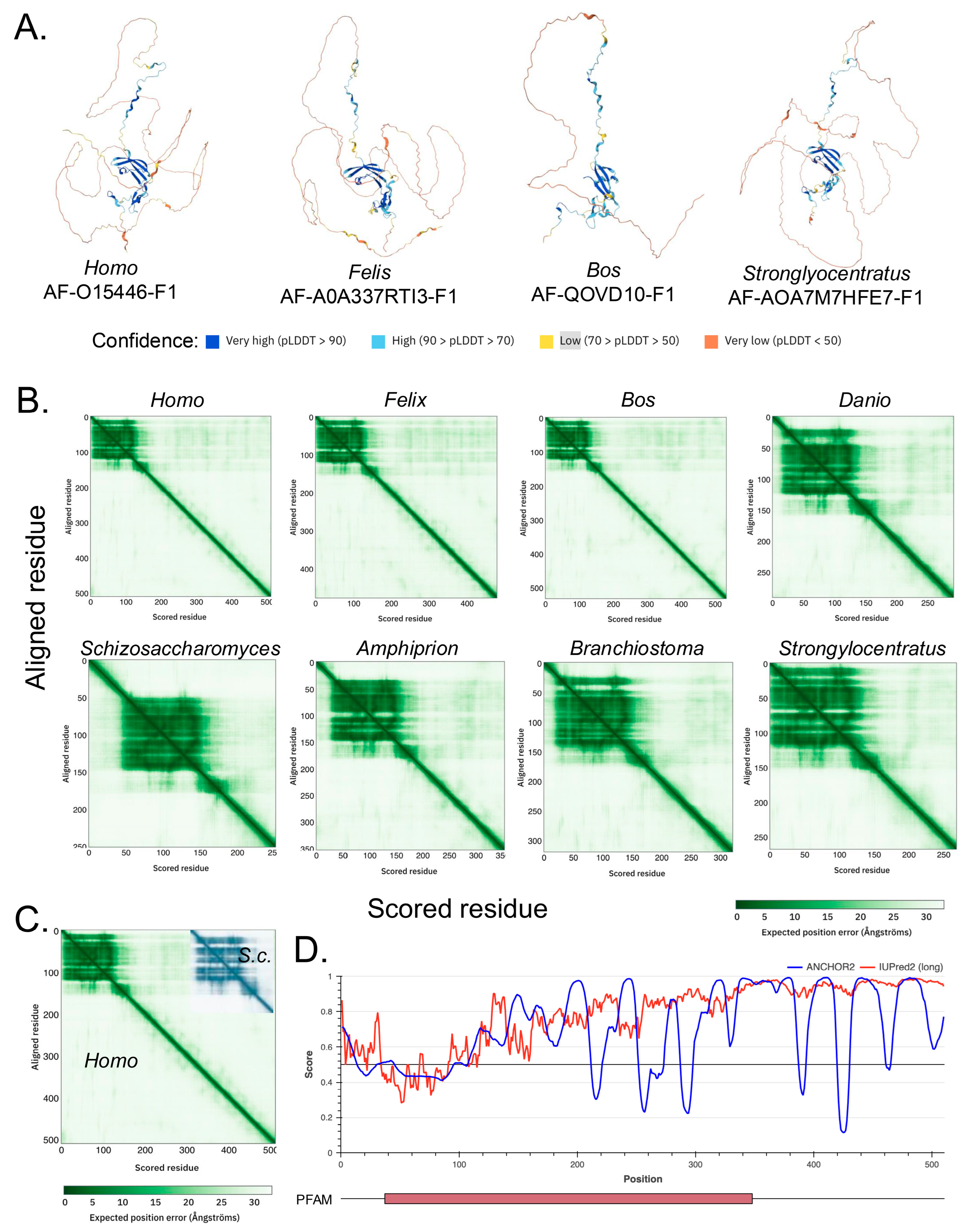
3.1. Structural Similarities and Evolutionary Trends in the A34 CTD
3.2. Repeated Sequences in the A34 CTD
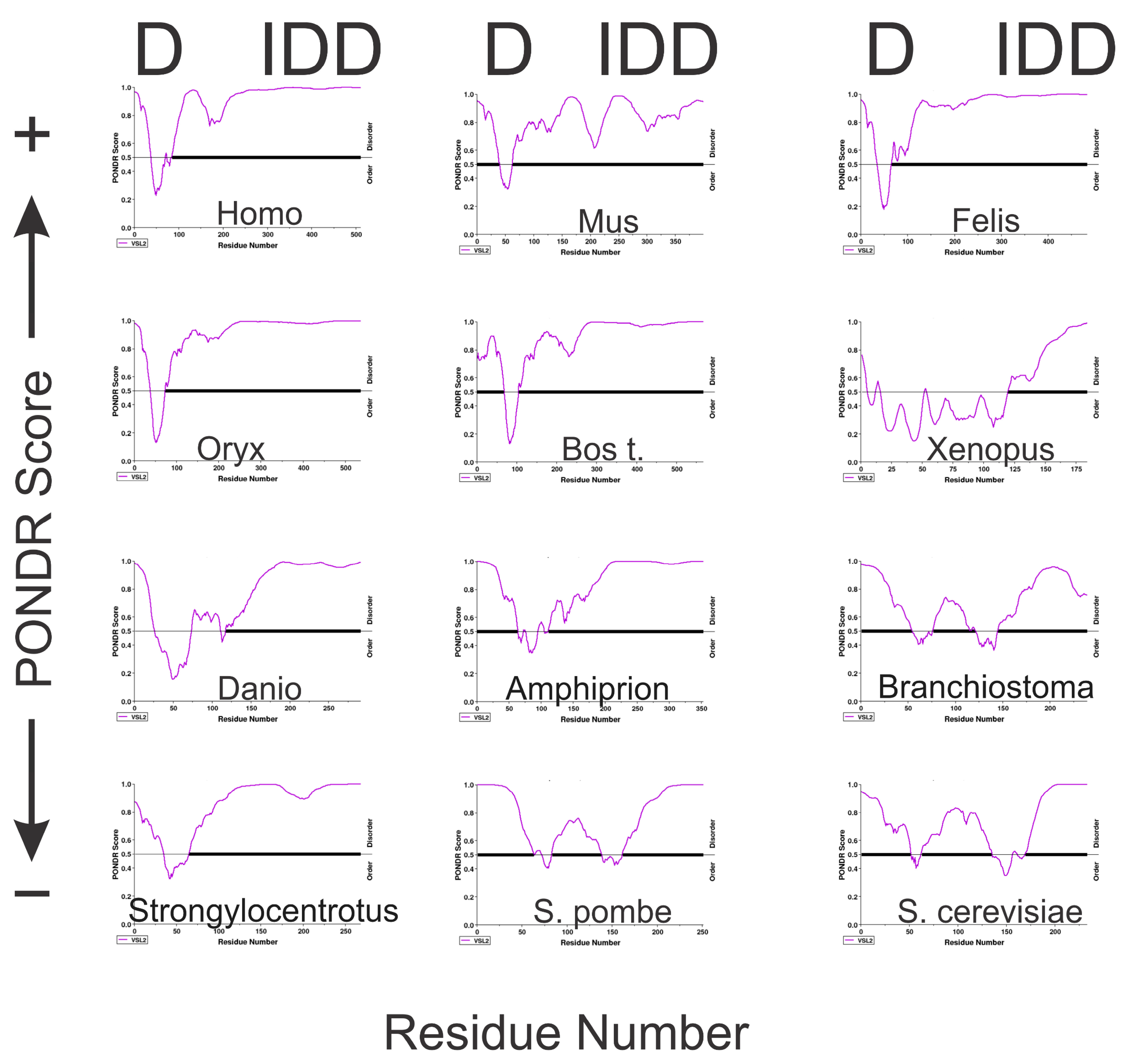
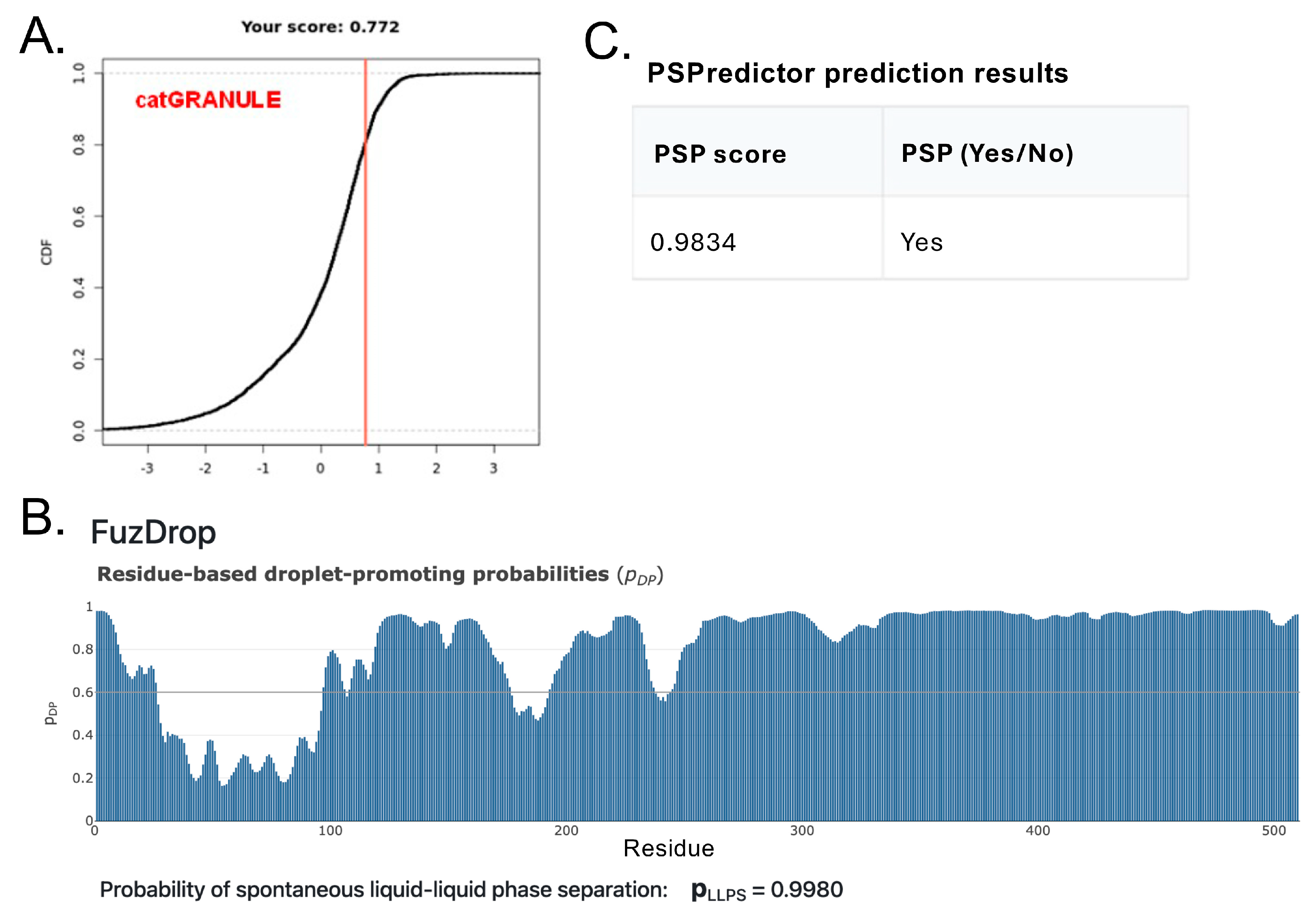
3.3. Assessing the Intrinsic Disorder and LLPS Propensity of the A34 CTD
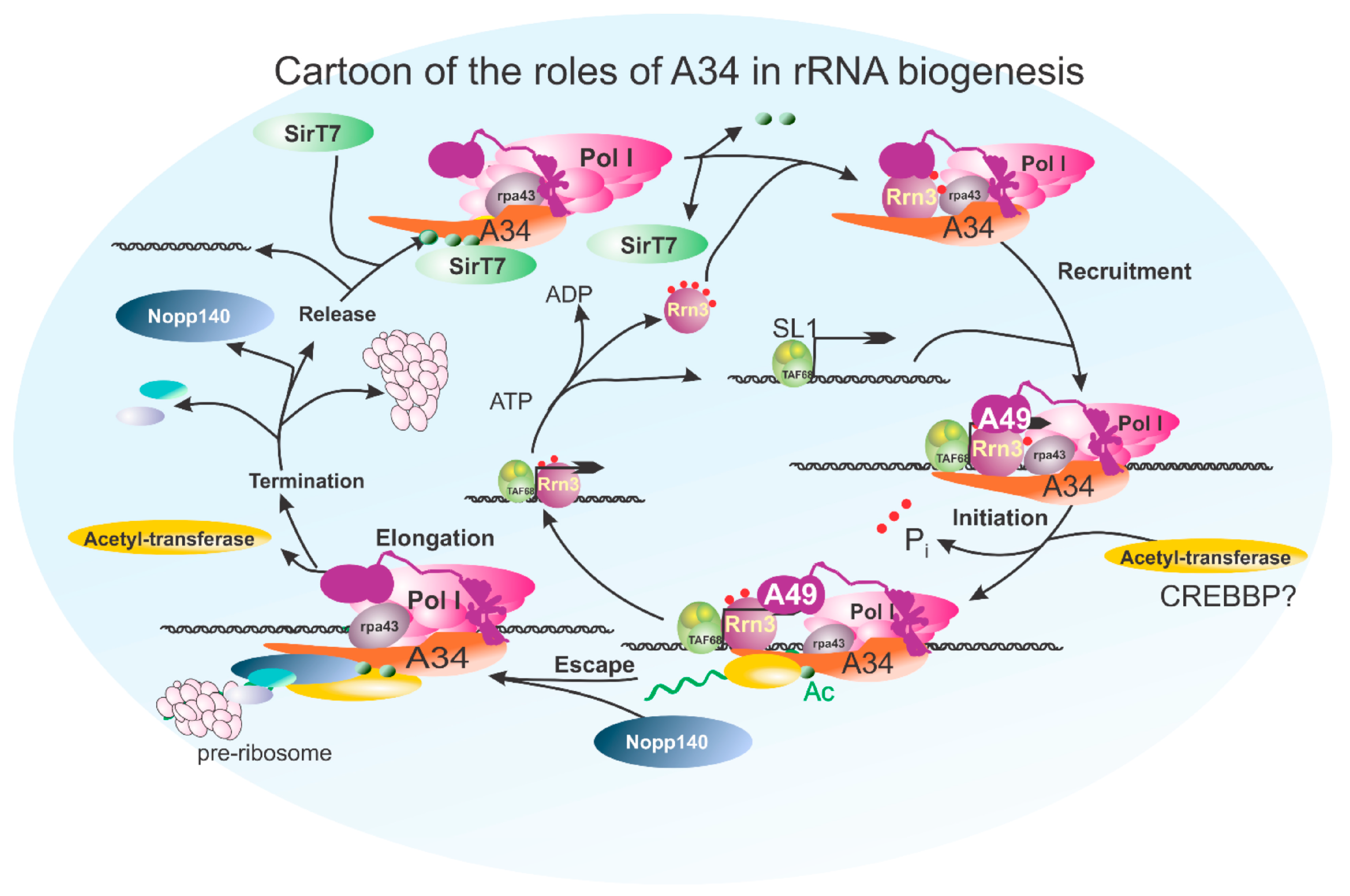
3.4. Post-Translational Modifications of the A34 CTD
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hannan, K.M.; Hannan, R.D.; Rothblum, L.I. Transcription by RNA polymerase I. Front. Biosci. A J. Virtual Libr. 1998, 3, d376–d398. [Google Scholar]
- Daiß, J.L.; Pilsl, M.; Straub, K.; Bleckmann, A.; Höcherl, M.; Heiss, F.B.; Abascal-Palacios, G.; Ramsay, E.P.; Tlučková, K.; Mars, J.C.; et al. The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans. Life Sci. Alliance 2022, 5, e202201568. [Google Scholar] [CrossRef] [PubMed]
- Misiaszek, A.D.; Girbig, M.; Grötsch, H.; Baudin, F.; Murciano, B.; Lafita, A.; Müller, C.W. Cryo-EM structures of human RNA polymerase I. Nat. Struct. Mol. Biol. 2021, 28, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Huet, J.; Buhler, J.M.; Sentenac, A.; Fromageot, P. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl. Acad. Sci. USA 1975, 72, 3034–3038. [Google Scholar] [CrossRef]
- Huet, J.; Dezélée, S.; Iborra, F.; Buhler, J.M.; Sentenac, A.; Fromageot, P. Further characterization of yeast RNA polymerases. Effect of subunits removal. Biochimie 1976, 58, 71–80. [Google Scholar] [CrossRef]
- Kuhn, C.D.; Geiger, S.R.; Baumli, S.; Gartmann, M.; Gerber, J.; Jennebach, S.; Mielke, T.; Tschochner, H.; Beckmann, R.; Cramer, P. Functional architecture of RNA polymerase I. Cell 2007, 131, 1260–1272. [Google Scholar] [CrossRef]
- Geiger, S.R.; Lorenzen, K.; Schreieck, A.; Hanecker, P.; Kostrewa, D.; Heck, A.J.; Cramer, P. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell 2010, 39, 583–594. [Google Scholar] [CrossRef]
- Engel, C.; Sainsbury, S.; Cheung, A.C.; Kostrewa, D.; Cramer, P. RNA polymerase I structure and transcription regulation. Nature 2013, 502, 650–655. [Google Scholar] [CrossRef]
- Fernandez-Tornero, C.; Moreno-Morcillo, M.; Rashid, U.J.; Taylor, N.M.; Ruiz, F.M.; Gruene, T.; Legrand, P.; Steuerwald, U.; Muller, C.W. Crystal structure of the 14-subunit RNA polymerase I. Nature 2013, 502, 644–649. [Google Scholar] [CrossRef]
- Knutson, B.A.; McNamar, R.; Rothblum, L.I. Dynamics of the RNA polymerase I TFIIF/TFIIE-like subcomplex: A mini-review. Biochem. Soc. Trans. 2020, 48, 1917–1927. [Google Scholar] [CrossRef]
- Gadal, O.; Mariotte-Labarre, S.; Chedin, S.; Quemeneur, E.; Carles, C.; Sentenac, A.; Thuriaux, P. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell. Biol. 1997, 17, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- Beckouet, F.; Labarre-Mariotte, S.; Albert, B.; Imazawa, Y.; Werner, M.; Gadal, O.; Nogi, Y.; Thuriaux, P. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol. Cell. Biol. 2008, 28, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- McNamar, R.; Freeman, E.; Baylor, K.N.; Fakhouri, A.M.; Huang, S.; Knutson, B.A.; Rothblum, L.I. PAF49: An RNA Polymerase I subunit essential for rDNA transcription and stabilization of PAF53. J. Biol. Chem. 2023, 299, 104951. [Google Scholar] [CrossRef]
- Panov, K.I.; Panova, T.B.; Gadal, O.; Nishiyama, K.; Saito, T.; Russell, J.; Zomerdijk, J.C. RNA polymerase I-specific subunit CAST/hPAF49 has a role in the activation of transcription by upstream binding factor. Mol. Cell. Biol. 2006, 26, 5436–5448. [Google Scholar] [CrossRef]
- Albert, B.; Leger-Silvestre, I.; Normand, C.; Ostermaier, M.K.; Perez-Fernandez, J.; Panov, K.I.; Zomerdijk, J.C.; Schultz, P.; Gadal, O. RNA polymerase I-specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle. J. Cell Biol. 2011, 192, 277–293. [Google Scholar] [CrossRef]
- Mangan, H.; Gailin, M.O.; McStay, B. Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 2017, 284, 3977–3985. [Google Scholar] [CrossRef]
- Feng, Z.; Chen, X.; Wu, X.; Zhang, M. Formation of biological condensates via phase separation: Characteristics, analytical methods, and physiological implications. J. Biol. Chem. 2019, 294, 14823–14835. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Erdős, G.; Dosztányi, Z. Analyzing Protein Disorder with IUPred2A. Curr. Protoc. Bioinform. 2020, 70, e99. [Google Scholar] [CrossRef]
- Linding, R.; Russell, R.B.; Neduva, V.; Gibson, T.J. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003, 31, 3701–3708. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; DiMaio, F.; Wang, R.Y.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-resolution comparative modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Chang, K.T.; Guo, J.; di Ronza, A.; Sardiello, M. Aminode: Identification of Evolutionary Constraints in the Human Proteome. Sci. Rep. 2018, 8, 1357. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, D.; Walsh, I.; Minervini, G.; Tosatto, S.C.E. FELLS: Fast estimator of latent local structure. Bioinformatics 2017, 33, 1889–1891. [Google Scholar] [CrossRef]
- Deng, W.; Wang, C.; Zhang, Y.; Xu, Y.; Zhang, S.; Liu, Z.; Xue, Y. GPS-PAIL: Prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci. Rep. 2016, 6, 39787. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Bolognesi, B.; Lorenzo Gotor, N.; Dhar, R.; Cirillo, D.; Baldrighi, M.; Tartaglia, G.G.; Lehner, B. A Concentration-Dependent Liquid Phase Separation Can Cause Toxicity upon Increased Protein Expression. Cell Rep. 2016, 16, 222–231. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Sequence determinants of the aggregation of proteins within condensates generated by liquid-liquid phase separation. J. Mol. Biol. 2022, 434, 167201. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Sun, T.; Li, Q.; Xu, Y.; Zhang, Z.; Lai, L.; Pei, J. Prediction of liquid-liquid phase separating proteins using machine learning. BMC Bioinform. 2022, 23, 72. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hou, C.; Wang, L.; Yu, C.; Chen, T.; Shen, B.; Hou, Y.; Li, P.; Li, T. Screening membraneless organelle participants with machine-learning models that integrate multimodal features. Proc. Natl. Acad. Sci. USA 2022, 119, e2115369119. [Google Scholar] [CrossRef]
- He, Y.; Yan, C.; Fang, J.; Inouye, C.; Tjian, R.; Ivanov, I.; Nogales, E. Near-atomic resolution visualization of human transcription promoter opening. Nature 2016, 533, 359–365. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Rosonina, E.; Blencowe, B.J. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA 2004, 10, 581–589. [Google Scholar] [CrossRef]
- Hsin, J.P.; Manley, J.L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012, 26, 2119–2137. [Google Scholar] [CrossRef]
- Portz, B.; Shorter, J. Switching Condensates: The CTD Code Goes Liquid. Trends Biochem. Sci. 2019, 45, 1–3. [Google Scholar] [CrossRef]
- Quintero-Cadena, P.; Lenstra, T.L.; Sternberg, P.W. RNA Pol II Length and Disorder Enable Cooperative Scaling of Transcriptional Bursting. Mol. Cell 2020, 79, 207–220. [Google Scholar] [CrossRef]
- Tellier, M.; Zaborowska, J.; Caizzi, L.; Mohammad, E.; Velychko, T.; Schwalb, B.; Ferrer-Vicens, I.; Blears, D.; Nojima, T.; Cramer, P.; et al. CDK12 globally stimulates RNA polymerase II transcription elongation and carboxyl-terminal domain phosphorylation. Nucleic Acids Res. 2020, 48, 7712–7727. [Google Scholar] [CrossRef]
- Babu, M.; Favretto, F.; Rankovic, M.; Zweckstetter, M. Peptidyl Prolyl Isomerase A Modulates the Liquid-Liquid Phase Separation of Proline-Rich IDPs. J. Am. Chem. Soc. 2022, 144, 16157–16163. [Google Scholar] [CrossRef] [PubMed]
- Mateos, B.; Conrad-Billroth, C.; Schiavina, M.; Beier, A.; Kontaxis, G.; Konrat, R.; Felli, I.C.; Pierattelli, R. The Ambivalent Role of Proline Residues in an Intrinsically Disordered Protein: From Disorder Promoters to Compaction Facilitators. J. Mol. Biol. 2020, 432, 3093–3111. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.X.; Kalmar, L.; Tompa, P.; Han, K.H.; Selenko, P.; Dunker, A.K.; Daughdrill, G.W.; Uversky, V.N. The alphabet of intrinsic disorder: I. Act like a Pro: On the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disord. Proteins 2013, 1, e24360. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Atencio, D.; Barnes, C.; DeFiglio, H.; Hanes, S.D. Multiple roles for the Ess1 prolyl isomerase in the RNA polymerase II transcription cycle. Mol. Cell. Biol. 2012, 32, 3594–3607. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yogesha, S.D.; Mayfield, J.E.; Zhang, Y. Cross-talk of phosphorylation and prolyl isomerization of the C-terminal domain of RNA Polymerase II. Molecules 2014, 19, 1481–1511. [Google Scholar] [CrossRef]
- Palacio, M.; Taatjes, D.J. Merging Established Mechanisms with New Insights: Condensates, Hubs, and the Regulation of RNA Polymerase II Transcription. J. Mol. Biol. 2022, 434, 167216. [Google Scholar] [CrossRef]
- Flores-Solis, D.; Lushpinskaia, I.P.; Polyansky, A.A.; Changiarath, A.; Boehning, M.; Mirkovic, M.; Walshe, J.; Pietrek, L.M.; Cramer, P.; Stelzl, L.S.; et al. Driving forces behind phase separation of the carboxy-terminal domain of RNA polymerase II. Nat. Commun. 2023, 14, 5979. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Mokin, Y.I.; Gavrilova, A.A.; Fefilova, A.S.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N.; Fonin, A.V. Nucleolar- and Nuclear-Stress-Induced Membrane-Less Organelles: A Proteome Analysis through the Prism of Liquid-Liquid Phase Separation. Int. J. Mol. Sci. 2023, 24, 11007. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2020, 22, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Hatos, A.; Tosatto, S.C.E.; Vendruscolo, M.; Fuxreiter, M. FuzDrop on AlphaFold: Visualizing the sequence-dependent propensity of liquid-liquid phase separation and aggregation of proteins. Nucleic Acids Res. 2022, 50, W337–W344. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Zhang, Y.; Qi, Y.; Zhang, Z. Evaluation of sequence-based predictors for phase-separating protein. Brief. Bioinform. 2023, 24, bbad213. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Olson, M.O.; Jones, C.; Busch, H. Nucleolar phosphoproteins of normal rat liver and Novikoff hepatoma ascites cells. Cancer Res. 1975, 35, 1470–1475. [Google Scholar] [PubMed]
- Olson, M.O.J. The Nucleolus; Landes Bioscience/Eurekah.Com; Kluwer Academic/Plenum Publishers: Georgetown, TX, USA; New York, NY, USA, 2004; 347p. [Google Scholar]
- Penrod, Y.; Rothblum, K.; Cavanaugh, A.; Rothblum, L.I. Regulation of the association of the PAF53/PAF49 heterodimer with RNA polymerase I. Gene 2015, 556, 61–67. [Google Scholar] [CrossRef]
- Cavanaugh, A.H.; Evans, A.; Rothblum, L.I. Mammalian Rrn3 is required for the formation of a transcription competent preinitiation complex containing RNA polymerase I. Gene Expr. 2008, 14, 131–147. [Google Scholar]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006, 20, 1075–1080. [Google Scholar] [CrossRef]
- Grob, A.; Roussel, P.; Wright, J.E.; McStay, B.; Hernandez-Verdun, D.; Sirri, V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 2009, 122, 489–498. [Google Scholar] [CrossRef]
- Chen, S.; Seiler, J.; Santiago-Reichelt, M.; Felbel, K.; Grummt, I.; Voit, R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell 2013, 52, 303–313. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Greco, T.M.; Cristea, I.M. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol. Cell. Proteom. 2014, 13, 73–83. [Google Scholar] [CrossRef]
- Sirri, V.; Grob, A.; Berthelet, J.; Jourdan, N.; Roussel, P. Sirtuin 7 promotes 45S pre-rRNA cleavage at site 2 and determines the processing pathway. J. Cell Sci. 2019, 132, jcs228601. [Google Scholar] [CrossRef]




| Gene | Protein | Aliases |
|---|---|---|
| RPA34/POLR1G | DNA-directed RNA polymerase I subunit RPA34 | A34, A34.5, PAF49, RNA polymerase I-associated factor PAF49, Antisense to ERCC-1 protein (ASE1), CD3-epsilon-associated protein (CD3EAP) |
| RPA49/POLR1E | DNA-directed RNA polymerase I subunit RPA49 | A49, PAF53, DNA-directed RNA polymerase I subunit E, RNA polymerase I-associated factor 1, RNA polymerase I-associated factor 53 |
| Organism | Pol II CTD Length | A34 CTD Length |
|---|---|---|
| Yeast | ~182 amino acids | 80 amino acids |
| Human | ~470 amino acids | 350 amino acids |
| Organism | Repeat Number |
|---|---|
| Homo s. | 9 |
| Felix c. | 8 |
| Mus m. | 7 |
| Danio r. | 4 |
| Strongylocentratus p. | 3 |
| Drosophila m. | 3 |
| Schizzosacharomyces p. | 3 |
| Saccharomyces c. | 3 |
| Nucleolar Protein | Percent Disordered |
|---|---|
| NOPP140 | 91 |
| PAF49 | 66.8 |
| Nucleolin (C23) | 64.9 |
| GAR1 | 59.5 |
| Nucleophosmin (B23) | 53.7 |
| NOP10 | 39.1 |
| Fibrillarin | 33.6 |
| NOP56 | 26.3 |
| Dyskerin | 24.3 |
| NHP2 | 15.7 |
| NHP2l1 | 6.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knutson, B.A.; Rothblum, L.I. Evolutionary and Structural Insights into the RNA Polymerase I A34 Protein Family: A Focus on Intrinsic Disorder and Phase Separation. Genes 2025, 16, 61. https://doi.org/10.3390/genes16010061
Knutson BA, Rothblum LI. Evolutionary and Structural Insights into the RNA Polymerase I A34 Protein Family: A Focus on Intrinsic Disorder and Phase Separation. Genes. 2025; 16(1):61. https://doi.org/10.3390/genes16010061
Chicago/Turabian StyleKnutson, Bruce A., and Lawrence I. Rothblum. 2025. "Evolutionary and Structural Insights into the RNA Polymerase I A34 Protein Family: A Focus on Intrinsic Disorder and Phase Separation" Genes 16, no. 1: 61. https://doi.org/10.3390/genes16010061
APA StyleKnutson, B. A., & Rothblum, L. I. (2025). Evolutionary and Structural Insights into the RNA Polymerase I A34 Protein Family: A Focus on Intrinsic Disorder and Phase Separation. Genes, 16(1), 61. https://doi.org/10.3390/genes16010061






