Abstract
Microgravity exposure induces a cephalad fluid shift and an overall reduction in physical activity levels which can lead to cardiovascular deconditioning in the absence of countermeasures. Future spaceflight missions will expose crew to extended periods of microgravity among other stressors, the effects of which on cardiovascular health are not fully known. In this study, we determined cardiac responses to extended microgravity exposure using the rat hindlimb unloading (HU) model. We hypothesized that exposure to prolonged simulated microgravity and subsequent recovery would lead to increased oxidative damage and altered expression of genes involved in the oxidative response. To test this hypothesis, we examined hearts of male (three and nine months of age) and female (3 months of age) Long–Evans rats that underwent HU for various durations up to 90 days and reambulated up to 90 days post-HU. Results indicate sex-dependent changes in oxidative damage marker 8-hydroxydeoxyguanosine (8-OHdG) and antioxidant gene expression in left ventricular tissue. Three-month-old females displayed elevated 8-OHdG levels after 14 days of HU while age-matched males did not. In nine-month-old males, there were no differences in 8-OHdG levels between HU and normally loaded control males at any of the timepoints tested following HU. RNAseq analysis of left ventricular tissue from nine-month-old males after 14 days of HU revealed upregulation of pathways involved in pro-inflammatory signaling, immune cell activation and differential expression of genes associated with cardiovascular disease progression. Taken together, these findings provide a rationale for targeting antioxidant and immune pathways and that sex differences should be taken into account in the development of countermeasures to maintain cardiovascular health in space.
1. Introduction
Spaceflight leads to cardiovascular deconditioning in the absence of mitigation strategies. Cardiovascular changes in response to spaceflight are attributed to altered microgravity levels and the ensuing cephalad fluid shift [1]. Overall reductions in physical activity and other factors such as nutritional changes, elevated CO2 levels, and a demanding workload also may be contributing factors. Some of the reported cardiovascular changes caused by exposure to the spaceflight environment include reductions in left ventricular mass [2,3], transient atrial distension [4], and hypovolemia [5]. Stagnant or retrograde venous flow as well as thrombosis have been observed in crew during flight [6]. Heart rhythm disturbances postflight also have been reported [7]. Orthostatic intolerance [8] and stiffer carotid arteries [9] have also been observed in crew upon return to Earth. Crew on exploration-class missions (e.g., trip to Mars) will experience prolonged periods of microgravity and exposure to low doses of space radiation given current spacecraft shielding designs. Therefore, it is important to understand the cardiovascular responses to long-duration spaceflight to mitigate potential health risks.
The rodent hindlimb unloading (HU) model [10,11] has been used to gain insights into some aspects of human cardiovascular responses to microgravity and cephalad fluid shifts. HU for four weeks in rats resulted in decreased left ventricular mass and volume [12], consistent with findings from a number of bed rest studies [2,13,14,15,16] and from spaceflight [3]. Two weeks of HU in rats perturbs day–night blood pressure (BP) and heart rate regulation [17]. Compared to baseline (pre-HU) values, HU generally led to increased systolic and diastolic BP during both day and night cycles, whereas heart rate was increased during the day but not at night. Some of these HU findings could also have been confounded by other factors such as social isolation and restraint stress [17]. In addition, arrhythmias and aberrant intracellular Ca2+ signaling have been observed in mice that underwent HU (26–56 days) [18]. Changes in redox balance have also been reported. For example, hearts from male Sprague–Dawley rats that underwent two weeks of HU displayed increased levels of malondialdehyde, a lipid peroxidation marker [19].
Currently, there is limited information regarding the effects of long-duration spaceflight on the human cardiovascular system at time scales relevant to interplanetary missions (>2 years). In addition, the effects of age, sex, duration of exposure, and long-term recovery on cardiovascular outcomes are not well established. Hence, the goals of our study are to (1) determine the impact of sex, age, duration of exposure, and recovery on oxidative stress responses to microgravity and (2) define underlying molecular mechanisms. We hypothesized that exposure to simulated microgravity and the ensuing recovery would lead to changes in redox balance and the expression of select genes including those that play a role in oxidative stress responses. To test this hypothesis, we examined hearts from male and female rats that underwent hindlimb unloading (HU) for various durations (up to 90 days) and those that underwent reambulation after 90 days of HU. To achieve this, we assessed DNA oxidative damage markers and select transcripts, and performed RNAseq analysis of cardiac tissue.
2. Materials and Methods
2.1. Animals and Experiment Design
Animal experiments were carried out in accordance with the recommendations of Guide for the Care and Use of Laboratory Animals, 8th edition. All animal experiments were conducted with prior approval from the UC Davis Institutional Animal Care and Use Committee (IACUC). Female and male Long–Evans rats (Taconic) were selected for this study. Animals were group-housed in standard vivarium cages until the onset of HU. A group of females and males (referred to as “young”) underwent HU at three months of age, while a group of retired male breeders (referred to as “older males”) began HU at nine months of age. Normally loaded (NL) controls were individually housed in standard vivarium cages. Animals were suspended onto the pulley apparatus via orthopedic traction tape as described previously [11]. NL controls were handled the same way as the HU animals except for the actual attachment of traction tape and tail suspension. HU was conducted for either 14 days or 90 days. A subset of nine-month-old (older) males that underwent HU for 90 days were then released from the suspension apparatus to reambulate normally (Reloading, Rel) for 90 days. All animals were supplied with standard rodent laboratory chow (Purina, St. Louis, MO, USA) and water ad libitum. Enrichment consisted of Diamond Twists paper sticks (Harlan, Indianapolis, IN, USA) and I-Chews (Newco, Hayward, CA, USA) and replenished as needed. Room temperature was maintained within a range of 20–26 °C with a 12-h light and 12-h dark cycle. After the designated duration of HU, animals were euthanized via isoflurane overdose followed by exsanguination and decapitation. Tissues were collected shortly thereafter. Hearts were bisected, flash-frozen in liquid nitrogen, and stored at −80 °C until processing. Refer to Table S1 for the sample sizes of the experiment groups included in this study. Young male and female, 90 day HU, and recovery groups were not included in the study either due to insufficient sample sizes or unavailability of appropriately processed samples.
2.2. Processing of Frozen Heart Samples
Frozen hearts were retrieved from −80 °C storage and placed on a metal platform that was pre-chilled with liquid nitrogen. Left ventricular tissue was excised from the anterior to posterior ends (apex) of the heart. The excised left ventricular tissue was further cut into thirds (anterior, middle, and posterior). The anterior and posterior thirds were processed for DNA and RNA extraction, respectively. Samples were excluded from analyses if anatomical landmarks could not be conclusively identified for isolation of left ventricular tissue.
2.3. RNA Extraction from Cardiac Tissue
Total RNA was extracted from frozen left ventricular tissue using the RNeasy Fibrous Tissue Mini Kit (Qiagen, Germantown, MD, USA) according to manufacturer’s protocol. RNA concentration was determined using NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and purity was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Only samples showing an RNA integrity number (RIN) > 8 were used for RNA sequencing and quantitative polymerase chain reaction (qPCR).
2.4. cDNA Synthesis and qPCR
First-strand DNA was synthesized from total RNA as follows. A total of 450 ng of RNA was used for a 30 uL first strand synthesis. Briefly, 1.5 uL of random hexamers (Invitrogen, Waltham, MA, USA, Cat# N8080127), 1.5 μL dNTPs (Qiagen, Cat# 1005631), and 12 μL of water were mixed, incubated at 65 °C for 5 min, and immediately chilled on ice. A solution-containing 3 μL RNase-free water, 6 μL 5× first strand buffer (Invitrogen, Cat# Y02321), 3 μL DTT (Invitrogen, Cat# Y00147), and 1.5 μL RNase Out (Invitrogen, Cat# 100000840) was added to the abovementioned mix and then incubated at 37 °C for 2 min. Then 1.5 μL of Moloney Murine Leukemia Virus (MMLV) reverse transcriptase (200 U/μL, Invitrogen, Waltham, MA, USA, Cat# 28025013) was added to the resulting solution and incubated as follows: 25 °C for 10 min, 37 °C for 50 min, and 70 °C for 15 min. All incubation steps were performed using a TC-312 thermocycler (Techne, Vernon Hills, IL, USA). First-strand DNA was stored at −80 °C until use for qPCR using the Taqman system. Multiplex PCR reactions were set up to measure both genes of interest and housekeeping gene expression in the same well of a 384-well plate. A premade gene expression master mix (Taqman, LifeTechnologies, Waltham, MA, USA, Cat# 4369016) was utilized, and each reaction was completed in duplicate. FAM-labeled Taqman probes (LifeTechnologies, Waltham, MA, USA) were used for the following genes of interest: nuclear factor, erythroid 2-like 2 (Nfe2l2, Rn00582415_m1), superoxide dismutase 1 (Sod1, Rn00566938_m1), superoxide dismutase 2 (Sod2, Rn00690588_g1), and sirtuin 1 (Sirt1, Rn01428096_m1). VIC-labeled ribosomal protein L19 (Rpl19, Rn00821265_g1) was used as housekeeping gene. qPCR was performed on a QuantStudio 6 Flex Real-Time PCR instrument (Applied Biosystems, Waltham, MA, USA). The relative gene expression levels for HU groups relative to age- and sex-matched controls were determined by the ΔΔCt method and reported as fold change.
2.5. 8-Hydroxydeoxyguanosine (8-OHdG) Assay
Total DNA was extracted from frozen left ventricular tissue using the DNeasy Blood and Tissue Kit (Qiagen) according to manufacturer’s protocol. Quantification of 8-OHdG was performed using OxiSelect™ Oxidative DNA Damage ELISA Kit (Cell Biolabs Inc., San Diego, CA, USA, Cat# STA-320) following manufacturer’s specifications. Enzyme reaction/absorbance was measured using a Spectra Max 250 (Molecular Devices, San Jose, CA, USA) and Softmax Pro 5 software.
2.6. RNA Sequencing and Bioinformatics Analysis
Poly(A)+-enriched cDNA libraries were prepared using the Illumina TruSeq RNA Library Prep kit (Illumina Inc., Hayward, CA, USA) and sequenced using Illumina NextSeq 550 (Illumina Inc., Hayward, CA, USA) instrument to generate 75 bp single-end reads. Sequencing data quality was checked using FastQC (version 0.11.5) software. Subsequently, reads were mapped to the rat reference genome (rn6) using STAR (version 2.6.0c). Then, a matrix of read counts per gene was generated using “featureCounts” from the Rsubread package (version 1.30.5). Genes with low expression values were filtered from downstream analysis by requiring more than 5 reads in at least 6 samples for each gene. Subsequently, between-sample normalization was performed using EDASeq (version 2.16.0). One sample from each group had to be excluded from analysis due to technical issues. Hence, 6 samples per group were used for the succeeding analysis. RUVseq (version 1.16.0) [RUVs with k = 4] was used to estimate the factors of unwanted variation. Genes differentially expressed between HU and control samples were identified using DESeq2 (version 1.22.2), controlling for factors of unwanted variation. Hierarchical clustering analysis was performed and heat maps generated using Morpheus, with Pearson method selected [20]. RStudio analysis software (Version 1.1.419), together with ggplot2 (version 3.1.1), ggrepel (version 0.8.0), and Bioconductor (DESeq2 version 1.22.2) packages, were used to obtain principal component analysis plots. For gene enrichment analysis, differentially expressed genes meeting a log2 fold change cut-off of >0.3 or <−0.3 and an adjusted p-value < 0.05 were input into Toppfun [21,22]. Toppfun was used to assess functional enrichment of DEGs based on ontologies, gene family, phenotype (human disease), and pharmacome (drug–gene associations). Of the 125 differentially expressed genes (DEGs), 121 matched the contents of the Toppfun database and were therefore included in the list for gene enrichment analysis. The DEGs that had no match in the Toppfun database were Clec4a1, Lilrb3l, Fcna, and Mill1. Gene enrichment analysis was performed at a false discovery rate (FDR, Benjamini Hochberg) of <0.05 and a gene limit of 2 or greater. GO Terms were plotted for visualization using the matplotlib.pyplot, seaborn, and pandas packages run in Google Colaboratory (Python ver 3.10).
2.7. Statistical Analysis
Equivalence of variance was first evaluated by Levene’s test. Once equal variance was confirmed, a t-test was performed to compare HU and matched control groups. This study involved an extended sampling period (3 years). To minimize batch effects, we compared HU and the matched controls for any given sex and timepoint since these groups were collected within the same period. Specifically, any HU group and its normally loaded control group (e.g., young male 14 day HU and NL groups) were euthanized on the same day or a day apart. Comparisons were not made across timepoints, sexes, or ages. Statistical analyses were performed using JMP software version 13.1.0 (SAS Institute Inc., Cary, NC, USA). Data shown are mean +/− S.D.
3. Results
In this study, we made use of the rodent HU model to gain insight on the effects of microgravity exposure on molecular signatures of the heart. With the exception of the 14-day HU young females, all HU groups had lower body weights versus age- and sex-matched controls (Figure 1), consistent with previous reports [23,24,25]. Weight loss in HU groups of older males was transient, with body weights of NL and HU groups being comparable after 90 days of reambulation. Due to the modest decrease in body weight (10–13%), we do not expect weight loss to be a major confounding factor in other experimental outcomes.
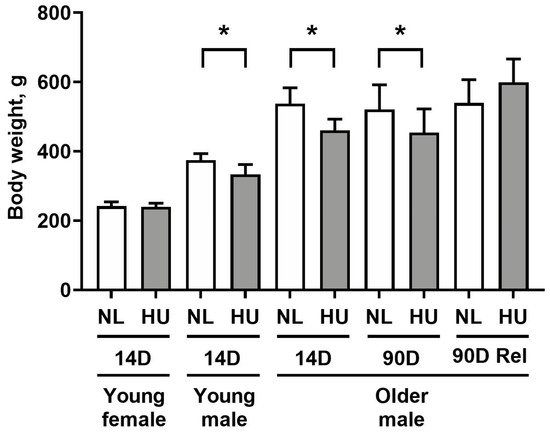
Figure 1.
Body weights of animals included in this study. NL: normally loaded; HU: hindlimb unloading; Rel: reambulated (reloading). The HU group was compared to their age- and sex-matched NL controls by Student’s t-test. Sample sizes: N = 5–8/group for all groups except for 90D older males, where N = 3. Values depicted are means and standard deviation. * Significant at p < 0.05 by Student’s t-test.
To determine whether simulated microgravity leads to alterations in redox balance in cardiac tissue, we measured levels of 8-hydroxydeoxyguanosine (8-OHdG) in left ventricular tissue (Figure 2). 8-OHdG is a marker for oxidative damage to DNA. Young females after 14 days of HU displayed a modest yet statistically significant increase in 8-OHdG levels (33.2%) compared to age- and sex-matched controls. At 14 days of HU, no differences were observed between HU and control groups in young and older males. Older males at 90 days of HU did not exhibit any significant difference in 8-OHdG levels versus corresponding controls. We further hypothesized that reloading will lead to increased oxidative damage to hearts due to altered fluid distribution and increased functional demand on the heart. Hence, we examined 8-OHdG levels at an earlier timepoint of reambulation (7 days) where we found no differences in 8-OHdG levels in older males that underwent normal reambulation after 90 days of HU relative to controls.
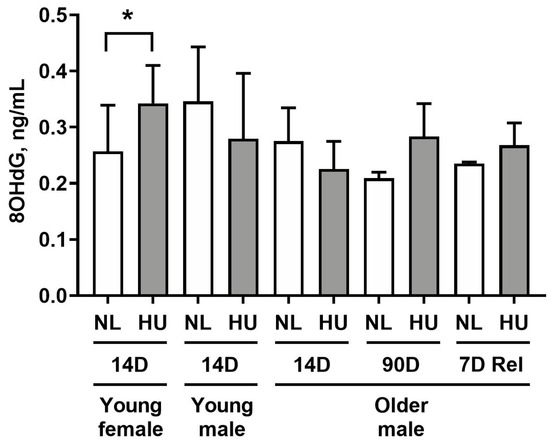
Figure 2.
8-hydroxydeoxyguanosine (8-OHdG) levels in left ventricular tissue as measured by ELISA. NL: normally loaded control, HU: hindlimb unloading, 7D Rel: 90D HU + 7D reloading and normally loaded control (NL). The HU groups were compared to their age- and sex-matched NL controls by Student’s t-test. Sample sizes: N = 5–8/group for all groups except for 90D older males, where N = 3. Values depicted are means and standard deviation. * Significant at p < 0.05 by Student’s t-test.
To further assess any changes in key molecular determinants of redox balance, we measured gene expression levels of select antioxidant genes (Sod1 and Sod2) as well as a master transcriptional regulator of the antioxidant response, Nfe2l2 (Figure 3A–C). In addition, we examined the gene expression of Sirt1, a marker of cellular senescence and oxidative stress also known to impact longevity [26,27] (Figure 3D). Compared to sex- and age-matched controls, young females that underwent 14 days of HU did not show any changes in gene expression for any of the four genes tested. Young males at the same timepoint showed a modest downregulation of Nfe2l2 (Figure 3A) as well as upregulation of Sod1 and Sod2 (Figure 3B and 3C respectively), but no change in Sirt1 expression (Figure 3D). In older males, Nfe2l2 was downregulated at 14 days of HU, while there were no changes in Sod1, Sod2, or Sirt1 expression. The downregulation of Nfe2l2 expression in older males persisted at 90 days of HU. After a 90-day reambulation period, this pattern was reversed, with HU groups showing upregulated expression of Nfe2l2 and Sirt1 relative to controls.
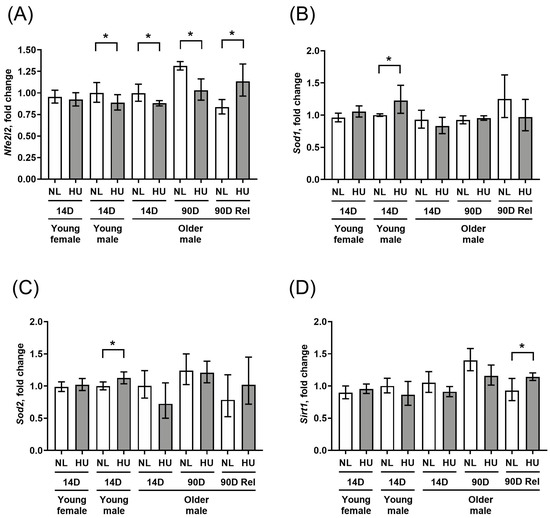
Figure 3.
Transcript levels of (A) Nfe2l2, (B) Sod1, (C) Sod2, and (D) Sirt1 in left ventricular wall as measured by qPCR. Values depicted are mean fold changes relative to young male control at 14 days of treatment as determined by the ΔΔCt method. Errors bars show upper and lower ranges. NL: normally loaded control, HU: hindlimb unloading, 90D Rel: 90D HU + 90D reloading and NL control. Sample sizes: N = 3–7/group. * Significant at p < 0.05 by Student’s t-test by comparing HU with age- and sex-matched NL control.
We reasoned that identifying the molecular signature of the early adaptation of cardiac tissue to HU will allow us to link early changes in gene expression and biological pathways to later cardiac outcomes. We therefore conducted transcriptomic analysis (RNAseq) of tissue from the left ventricle in older (nine-month-old) males and their corresponding controls. We selected nine-month-old males for analysis (samples were not available for females of similar age) over the other three-month-old groups. Principal component analysis (PCA) and hierarchical clustering generally indicated clustering by treatment group (Figure 4 and Figure 5 respectively). A total of 125 genes were differentially expressed in the HU group relative to controls. Of these, 48 were upregulated and 77 were downregulated (Table 1). The top 10 upregulated and downregulated genes included a number of immune-related genes such as Tlr8 and the macrophage/monocyte markers CD68 and CD163 as well as two circadian clock-related genes, Per3 and Arntl (Figure 6). Fifteen of these DEG’s were cluster-of-differentiation (CD) molecules belonging to the C-type lectin domain family, while 6 were LY6/PLAUR domain-containing members of the complement gene family (Table 2). Nfe2l2 did not appear as a downregulated gene in the 14D HU hearts of older males based on RNAseq despite the observed decrease (although modest) in transcript levels by qPCR analysis (Figure 3A, Table 1). This is likely due to differences in the normalization methods for the two assays.
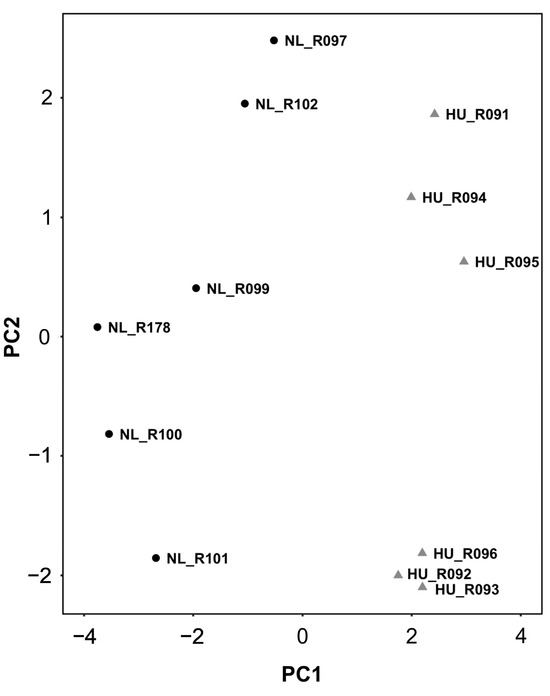
Figure 4.
PCA plot of transcriptomic data from older males that underwent 14 days of HU (gray triangles) and corresponding NL controls (black circles). Animal ID is indicated by the letter “R” succeeded by numbers.
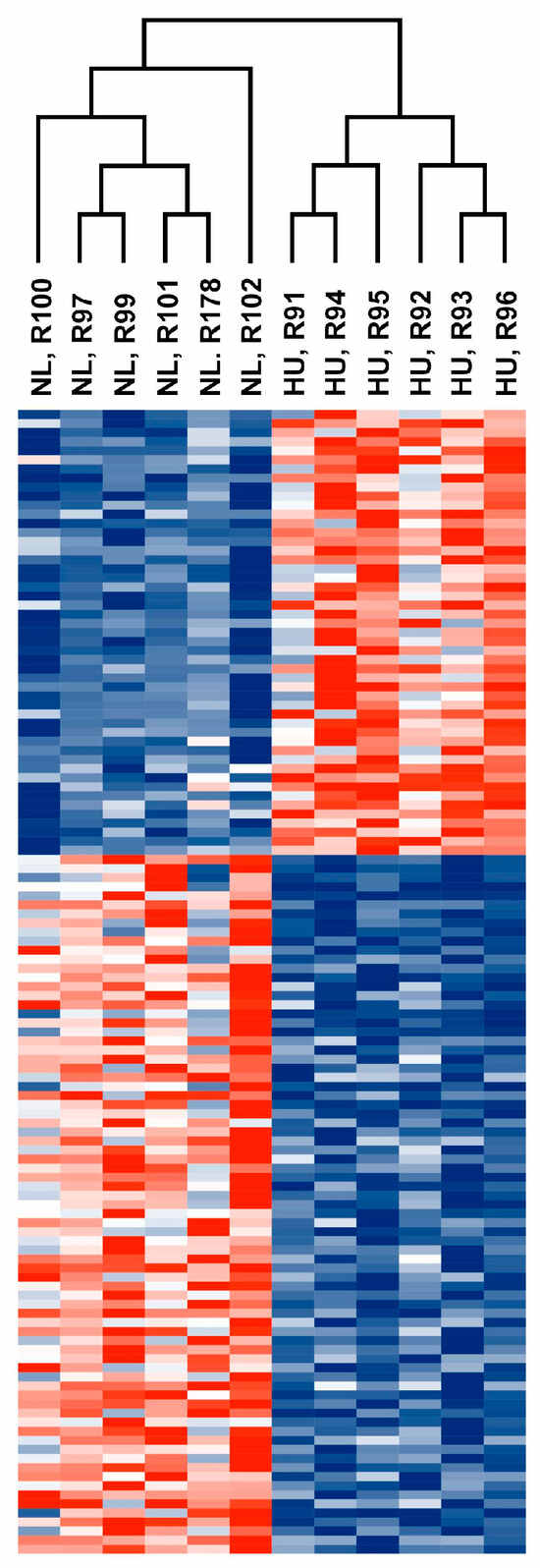
Figure 5.
Heatmap showing normalized counts of differentially expressed genes from older male 14D NL and HU groups. Each cell corresponds to a gene. Red: Upregulated in 14D HU relative to NL group. Blue: Downregulated in 14D HU relative to NL group. Magnitude of upregulation or downregulation is proportional to the intensity of red or blue. Deepest red: most upregulated; deepest blue: most downregulated.

Table 1.
Differentially expressed genes (DEGs) in older male 14D HU group relative to age- and sex- matched NL control group. DEGs were selected using an adj p-value < 0.05 at a log2 fold change cut-off > 0.3 or <−0.3. Total DEGs: 125 (48 upregulated and 77 downregulated).
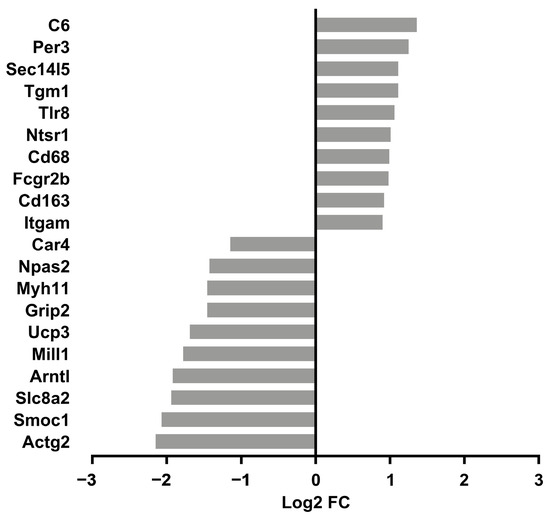
Figure 6.
Top 10 upregulated and downregulated genes in older male 14D HU relative to NL groups. Log2 FC: Log2 fold change. Refer to Table 1 for full list of DEGs.

Table 2.
Top gene families represented within DEGs.
Gene enrichment analysis of DEGs that met the FDR threshold revealed a predominance of immune-related processes including macrophage activation and complement activation. Further, circadian signaling, vesicle-mediated transport, and redox signaling were differentially enriched (Figure 7; refer to Table S2 for full list). Gene enrichment analysis for disease phenotypes using Toppfun [21,22] also revealed enrichment of a subset of DEGs linked to cardiovascular disease (CVD), immune dysfunction, neurovascular disease, cancer, metabolic disease, and sleep disorders. Cardiovascular pathologies represented include hypertensive disease, myocardial infarction, and heart failure. Immune disorders include complement deficiency and the autoimmune disease lupus erythematosus (Table 3; refer to Table S3 for full list). Consistent with the human disease phenotype findings, 24 DEGs were associated with the anti-bradycardia drug isoproterenol, a non-selective β adrenergic receptor agonist. In addition, a number of DEGs were associated with anti-hypertensive drugs losartan and valsartan, both angiotensin receptor blockers. A subset of DEGs also were associated with simvastatin (anticholesterolemic), doxorubicin (cancer treatment), rosiglitazone (antidiabetic), and melatonin (sleeping aid) (Table 4; refer to Table S4 for full list).
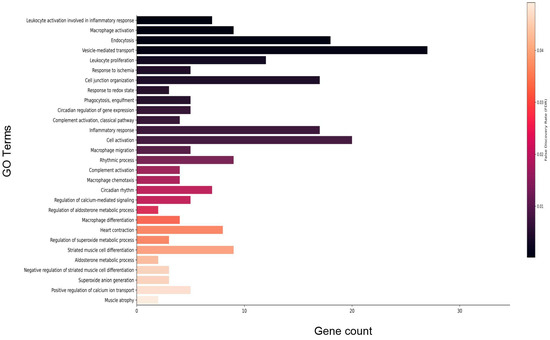
Figure 7.
Select enriched gene ontology (GO) terms for biological processes in older male 14D HU group relative to NL group. Gene count refers to the number of DEGs that matched the GO term. The vertical bar represents the color scale of the FDR with black representing the lowest FDR. Refer to Table S2 for the full list of GO terms.

Table 3.
Select human diseases associated with a subset of DEGs in this study. The last column indicates the number of DEGs that overlap with the known set of genes associated with human disease. Refer to Table S3 for full list. CVD: Cardiovascular disease.

Table 4.
Select pharmacologicals associated with a subset of DEGs in this study. The list contains current and discontinued drugs. Refer to Table S4 for the full list of pharmacologicals and associated DEGs. CVD: cardiovascular disease, HIV: human immunodeficiency virus, MS: multiple sclerosis.
4. Discussion
The objective of this study was to determine the impact of simulated microgravity on redox signaling in the heart and the overall transcriptomic landscape. Our findings from a rat hindlimb unloading (HU) model for cephalad fluid shift and microgravity exposure suggest sex and age differences in oxidative stress responses. Young males displayed upregulation in the expression of the antioxidants Sod1 and Sod2, and a decrease in Nfe2l2 expression, with no changes in 8-OHdG levels. On the other hand, increased levels of the DNA oxidative marker 8-OHdG was observed in females that underwent 14 days of HU with no changes in the expression levels of the redox signaling-related genes examined. One interpretation of our findings is that during short-term HU, young males mitigate oxidative damage more effectively than young females. Alternatively, it is possible that the kinetics of the emergence and repair of oxidative damage are sexually dimorphic. It is also unclear whether sex differences in oxidative stress responses persist due to unavailability of samples at later timepoints. Further, short-term HU in older males does not appear to enhance oxidative damage. Although not statistically significant, there was a trend toward increased 8-OHdG in the older males that underwent 90 days of HU compared to controls. We cannot rule out the possibility that long-duration HU can increase oxidative damage in the hearts of older males due to the small sample size available to perform the 8-OHdG assay in these groups. At 14 days of HU, older males exhibited a downregulation of Nfe2l2 expression, while 8-OHdG levels were not different between HU and controls. The persistent downregulation of Nfe2l2 at this timepoint is consistent with the observed trend towards increased oxidative damage during prolonged HU. A 90 day reambulation period after prolonged HU of 90 days led to upregulated Nfe2l2 and Sirt1 expression. It is possible that upregulation of Nfe2l2 and Sirt1 may be one of the underlying mechanisms by which the heart can recover from extended durations of HU. We recognize the extended time period for the conduct of the HU experiments and varying sample sizes across groups as limitations of our study. Specifically, the interpretation of the results from the 8-OHdG and qPCR assays which were performed on multiple HU groups and their corresponding NL controls merits further study. Consistent with our findings, another group has reported that HU elicits an antioxidant response. In the hearts of 6-month-old mice, HU led to time-dependent increases in the oxidation of glutathione to glutathione disulfide, an indicator of oxidative damage [28]. Taken together, our findings and those of others support the use of antioxidant-based countermeasures for extended duration spaceflight.
RNAseq results indicate that 14 days of HU in older males leads to enrichment of processes involved in the immune response, including complement activation and inflammation (Figure 7, Table S2). Complement activation, a component of innate immunity, is a mechanism by which the host defends against microbial infections. Studies in the last decade have revealed a role for the complement system in recognizing damaged host cells and coordinating with other elements of the immune response to achieve resolution to injury [29,30,31]. Dysregulated complement responses have been linked to autoimmune disease [32] as well as poor outcomes in patients with cardiovascular disease [33,34,35] and related animal models [36,37]. Our results indicate upregulation of several complement genes. These include C1qa, C1qb, and C1qc, all components of the membrane attack complex, as well as C5ar1, the receptor for the complement anaphylatoxin 5a, which can stimulate immune cells in myocardial infarct models [29,36,37] (Table 1). The implication of upregulated complement response in cardiac tissue during simulated microgravity requires further study given the emerging role of the complement pathway in the development of cardiovascular disease [37,38]. Interestingly, in International Space Station (ISS) crew, elements of the complement response were found to be modestly increased within a week of return to Earth relative to pre-flight values [39]. However, the impact of actual flight on complement responses could not be evaluated, since no in-flight measures were reported in the said study.
The underlying bases for the observed upregulation of complement genes in HU hearts is unclear. In Earth-based cardiovascular disease, altered fluid distribution and pressure changes can give rise to injury of cardiac tissue and therefore may play a role in the upregulated complement response seen in this rodent model for microgravity-induced fluid shifts. Complement activation also can occur as a response to pathogens or their constituents. Hence, another possibility that requires further study is the role of gut permeability in promoting a complement response during simulated microgravity. HU has been reported to alter the gut microbiome, cause damage to intestinal villi, and compromise the structural integrity of tight junctions [40], which can lead to gut leakiness. Indeed, HU can cause a transient release of lipopolysaccharide (LPS) in circulation and changes in the distribution of neutrophils and lymphocytes, [41] as well as an increase in circulating levels of the potent pro-inflammatory cytokine interferon γ (IFNγ) [42]. Further, synthetic bacterial DNA has been shown to induce a robust inflammatory response in the heart of mice [43]. Consistent with a potential role of gut permeability, we found upregulation of CD14 in 14-day HU hearts. CD14 is a surface receptor expressed in circulating monocytes/macrophages, which binds to LPS to induce an immune response [44]. Other key genes involved in the inflammatory response were also upregulated after 14 days of HU, including CD163 and CD68. CD163 is a marker for activated macrophages. CD163+ macrophages promote angiogenesis, vessel permeability, and leukocyte infiltration [45] and are increased in hearts of simian immunodeficiency virus-infected monkeys [46]. Further, CD68 marks the monocyte/macrophage population. Both expression levels of CD68 and immunopositive cell counts increase in the hearts of mice exposed to spaceflight-relevant doses (<0.5 Gy) of radiation [47]. In addition, HU in mice increased hippocampal CD68 levels, which mark activated microglial populations [48,49]. Collectively, the upregulation of CD14, CD68, and CD163 expression would be consistent with macrophage infiltration in HU hearts. Humans in future deep space missions will experience extended periods of low-dose space radiation and microgravity. Hence, the impact of the predicted upregulation of macrophage activity on human cardiovascular health requires further study. Tlr8 also was upregulated in the hearts of 14-day HU animals. Accumulating evidence demonstrates the important role of TLR signaling in the development of cardiovascular disease [50,51], whereas Tlr8 transcripts increased in patients with enterovirus-induced dilated cardiomyopathy [52].
RNAseq results also suggest that HU in older males can perturb cardiac circadian clock signals. The circadian clock gene, Arntl (Bmal1), was the fourth most downregulated gene in the HU group, while Per3 was the most upregulated gene. Other circadian clock-related genes such as Per2 and Clock were upregulated and downregulated, respectively. There is a growing appreciation for the role of clock genes in cardiovascular disease. Cardiac-specific deletion of Arntl leads to congestive heart failure during aging, upregulation of oxidative stress-responsive genes in hearts and altered energy metabolism [53]. In addition, Per2 gene ablation worsens the inflammatory response to myocardial ischemia [54]. ISS crew experience circadian misalignment which leads to sleep–wake cycle disruption [55]. Bioinformatic analysis of tissues from rodent spaceflight and ground-based studies (e.g., skeletal muscle, adrenals, kidney, liver) consistently show that spaceflight and its analogs lead to altered expression of circadian clock-related genes including Per2, Arntl, and Clock [56]. Taken together, our findings and those of others show the importance of exploring the role of circadian clock signaling in spaceflight-induced tissue deficits.
RNAseq also revealed the enrichment of processes related to redox signaling, such as superoxide anion generation. Furthermore, 14 days of HU led to differential expression of a number of genes known to play a role in the defense against oxidative damage. In the current study, Clusterin (Clu), also known as Apolipoprotein J (ApoJ), was upregulated after 14 days of HU. Overexpression of Clu in rat ventricular cells has been reported to rescue angiotensin II-induced ROS production and apoptosis [57]. Ucp3, a member of the family of mitochondrial uncoupling proteins (UCPs), was downregulated at 14 days of HU. UCPs have a number of functions, such as dissipating the proton gradient in the mitochondria, which reduces ATP production, releasing energy as heat, and also play a role in the regulation of redox balance [58]. Compared to wild-type controls, the hearts of Ucp3 knockout mice maintained in thermoneutral conditions have reduced mitochondrial complex activities as well as increased levels of ROS and oxidative stress markers [59]. Thus, the downregulation of Ucp3 in the hearts of HU rats suggests that simulated microgravity alters redox signaling and energetic processes in the mitochondria, consistent with findings from spaceflight and analog studies of other tissues [60,61]. Regulator of Calcineurin 1 (Rcan1), which is downregulated in hearts after 14 days of HU, plays a role in NFAT/Calcineurin signaling and is implicated in CVD progression [62,63]. Deletion of Rcan1 exacerbates the effects of septic cardiomyopathy and increases cardiac mitochondrial injury, as shown by elevated ROS levels and loss of mitochondrial membrane potential [62]. Taken together, our findings are in support of the hypothesis that simulated microgravity leads to altered redox signaling and function in the heart. In addition, these results are consistent with our earlier findings that hearts of mice that have been in space for 15 days show altered expression of genes regulating redox balance [64]. Taken together, the overall transcriptomic signature in the left ventricle of older males at 14 days of HU (~1 human year) suggests activation of the immune system and pro-inflammatory signaling, altered expression of circadian clock genes, and induction of signals that promote ROS production together with upregulation of select cellular defenses to oxidative damage. Therefore, our model predicts that in the absence of any mitigation strategies, extended spaceflight (>a year) may upregulate pro-inflammatory signaling in cardiac tissue. Consistent with our hypothesis, simulated microgravity led to enhanced expression of genes involved in the inflammatory and oxidative stress responses. Therefore, mitigating excessive inflammation and ensuring optimum antioxidant defenses (e.g., via dietary supplementation) may be a useful approach to ensure long-term cardiac health in astronauts during deep space missions. The upregulation of complement activation genes in hearts of rodents exposed to HU needs further study to determine its underlying basis and long-term consequences for cardiac health. In addition, we have identified the molecular signature of cardiac tissue responses to HU. A subset of these differentially expressed genes could be tested as candidate circulating biomarkers for monitoring the trajectory of an astronaut’s response to spaceflight or predicting the need for medical intervention.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15080975/s1, Table S1: List of groups and sample sizes included in analyses, Table S2: Results of functional enrichment analysis using Toppfun, Table S3: Results of human disease phenotype analysis using Toppfun, Table S4: Results of drug-gene association analysis using Toppfun.
Author Contributions
Conceptualization, Y.S., R.K.G. and C.G.T.T.; methodology, C.G.T.T., A. Sebastian, G.G.L., D.A.G., M.T., N.R.H., M.A. and M.C.; formal analysis, C.G.T.T., A. Sebastian, M.T., S.S., N.R.H., M.A. and M.C.; resources, R.K.G., G.G.L., D.A.G. and C.A.F.; visualization, C.G.T.T.; writing—original draft preparation, C.G.T.T.; writing—review and editing, all authors; project administration, T.M.H.-H.; supervision, C.G.T.T., R.K.G., A. Schreurs and T.M.H.-H.; funding acquisition, R.K.G., Y.S., A. Schreurs and C.A.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from the NASA Human Research Program Human Health Countermeasures (HRP HHC) NNJ14ZSA001N-FLAGSHIP and NNJ13ZSA002N-FLAGSHIP (both to R.K.G.) and NNX13AD94G to C.A.F. A. Schreurs and M.T. were supported by fellowships from the NASA Space Biology Postdoctoral Program (NPP). Work by G.G.L., N.R.H., and A. Sebastian were performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
Institutional Review Board Statement
The animal study protocol (Protocol 17390) was approved by the Institutional Animal Care and Use Committee of the University of California at Davis on 24 January 2013 and renewed annually for the duration of the project.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Material.
Acknowledgments
We are grateful to April Ronca and other members of Ames SC Division for coordinating this NASA-funded tissue-sharing opportunity. We also thank Sungshin Choi and other members of the NASA Ames and UC Davis dissection teams for assistance in collecting rat tissue samples.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Norsk, P.; Asmar, A.; Damgaard, M.; Christensen, N.J. Fluid shifts, vasodilatation and ambulatory blood pressure reduction during long duration spaceflight. J. Physiol. 2015, 593, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Perhonen, M.A.; Franco, F.; Lane, L.D.; Buckey, J.C.; Blomqvist, C.G.; Zerwekh, J.E.; Peshock, R.M.; Weatherall, P.T.; Levine, B.D. Cardiac atrophy after bed rest and spaceflight. J. Appl. Physiol. 2001, 91, 645–653. [Google Scholar] [CrossRef]
- Summers, R.L.; Martin, D.S.; Meck, J.V.; Coleman, T.G. Mechanism of spaceflight-induced changes in left ventricular mass. Am. J. Cardiol. 2005, 95, 1128–1130. [Google Scholar] [CrossRef] [PubMed]
- Khine, H.W.; Steding-Ehrenborg, K.; Hastings, J.L.; Kowal, J.; Daniels, J.D.; Page, R.L.; Goldberger, J.J.; Ng, J.; Adams-Huet, B.; Bungo, M.W.; et al. Effects of Prolonged Spaceflight on Atrial Size, Atrial Electrophysiology, and Risk of Atrial Fibrillation. Circ. Arrhythm. Electrophysiol. 2018, 11, e005959. [Google Scholar] [CrossRef] [PubMed]
- Watenpaugh, D.E. Fluid volume control during short-term space flight and implications for human performance. J. Exp. Biol. 2001, 204, 3209–3215. [Google Scholar] [CrossRef]
- Marshall-Goebel, K.; Laurie, S.S.; Alferova, I.V.; Arbeille, P.; Aunon-Chancellor, S.M.; Ebert, D.J.; Lee, S.M.C.; Macias, B.R.; Martin, D.S.; Pattarini, J.M.; et al. Assessment of Jugular Venous Blood Flow Stasis and Thrombosis During Spaceflight. JAMA Netw. Open 2019, 2, e1915011. [Google Scholar] [CrossRef]
- Migeotte, P.F.; Prisk, G.K.; Paiva, M. Microgravity alters respiratory sinus arrhythmia and short-term heart rate variability in humans. Am. J. Physiol. Heart Circ. Physiol. 2003, 284, H1995–H2006. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.C.; Feiveson, A.H.; Stein, S.; Stenger, M.B.; Platts, S.H. Orthostatic Intolerance After ISS and Space Shuttle Missions. Aerosp. Med. Hum. Perform. 2015, 86, A54–A67. [Google Scholar] [CrossRef] [PubMed]
- Hughson, R.L.; Robertson, A.D.; Arbeille, P.; Shoemaker, J.K.; Rush, J.W.; Fraser, K.S.; Greaves, D.K. Increased postflight carotid artery stiffness and inflight insulin resistance resulting from 6-mo spaceflight in male and female astronauts. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H628–H638. [Google Scholar] [CrossRef]
- Globus, R.K.; Morey-Holton, E. Hindlimb unloading: Rodent analog for microgravity. J. Appl. Physiol. 2016, 120, 1196–1206. [Google Scholar] [CrossRef]
- Morey-Holton, E.R.; Globus, R.K. Hindlimb unloading rodent model: Technical aspects. J. Appl. Physiol. 2002, 92, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.X.; Zhang, L.F.; Shang, H.H.; Yu, Z.B.; Qian, Y.Q. Echocardiographic assessment of left ventricular structure and function after simulated weightlessness in rats. Space Med. Med. Eng. (Beijing) 1999, 12, 88–91. [Google Scholar]
- Levine, B.D.; Zuckerman, J.H.; Pawelczyk, J.A. Cardiac atrophy after bed-rest deconditioning: A nonneural mechanism for orthostatic intolerance. Circulation 1997, 96, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, T.A.; Levine, B.D.; Tillery, T.; Peshock, R.M.; Hastings, J.L.; Schneider, S.M.; Macias, B.R.; Biolo, G.; Hargens, A.R. Cardiac atrophy in women following bed rest. J. Appl. Physiol. 2007, 103, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Westby, C.M.; Martin, D.S.; Lee, S.M.; Stenger, M.B.; Platts, S.H. Left ventricular remodeling during and after 60 days of sedentary head-down bed rest. J. Appl. Physiol. 2016, 120, 956–964. [Google Scholar] [CrossRef]
- Shibata, S.; Perhonen, M.; Levine, B.D. Supine cycling plus volume loading prevent cardiovascular deconditioning during bed rest. J. Appl. Physiol. 2010, 108, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Tsvirkun, D.; Bourreau, J.; Mieuset, A.; Garo, F.; Vinogradova, O.; Larina, I.; Navasiolava, N.; Gauquelin-Koch, G.; Gharib, C.; Custaud, M.A. Contribution of social isolation, restraint, and hindlimb unloading to changes in hemodynamic parameters and motion activity in rats. PLoS ONE 2012, 7, e39923. [Google Scholar] [CrossRef] [PubMed]
- Respress, J.L.; Gershovich, P.M.; Wang, T.; Reynolds, J.O.; Skapura, D.G.; Sutton, J.P.; Miyake, C.Y.; Wehrens, X.H. Long-term simulated microgravity causes cardiac RyR2 phosphorylation and arrhythmias in mice. Int. J. Cardiol. 2014, 176, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P.; Soulsby, M.E.; Scott, J.L. Effects of aminoguanidine on tissue oxidative stress induced by hindlimb unloading in rats. Ann. Clin. Lab. Sci. 2009, 39, 64–70. [Google Scholar]
- Morpheus. Available online: https://software.broadinstitute.org/morpheus/ (accessed on 13 April 2019).
- Chen, J.; Aronow, B.J.; Jegga, A.G. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinform. 2009, 10, 73. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Ulanova, A.; Gritsyna, Y.; Salmov, N.; Lomonosova, Y.; Belova, S.; Nemirovskaya, T.; Shenkman, B.; Vikhlyantsev, I. Effect of L-Arginine on Titin Expression in Rat Soleus Muscle After Hindlimb Unloading. Front. Physiol. 2019, 10, 1221. [Google Scholar] [CrossRef]
- Marzuca-Nassr, G.N.; Fortes, M.A.S.; Guimaraes-Ferreira, L.; Murata, G.M.; Vitzel, K.F.; Vasconcelos, D.A.A.; Bassit, R.A.; Curi, R. Short-term creatine supplementation changes protein metabolism signaling in hindlimb suspension. Braz. J. Med. Biol. Res. 2019, 52, e8391. [Google Scholar] [CrossRef] [PubMed]
- Dehority, W.; Halloran, B.P.; Bikle, D.D.; Curren, T.; Kostenuik, P.J.; Wronski, T.J.; Shen, Y.; Rabkin, B.; Bouraoui, A.; Morey-Holton, E. Bone and hormonal changes induced by skeletal unloading in the mature male rat. Am. J. Physiol. 1999, 276, E62–E69. [Google Scholar] [CrossRef] [PubMed]
- North, B.J.; Rosenberg, M.A.; Jeganathan, K.B.; Hafner, A.V.; Michan, S.; Dai, J.; Baker, D.J.; Cen, Y.; Wu, L.E.; Sauve, A.A.; et al. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 2014, 33, 1438–1453. [Google Scholar] [CrossRef]
- Wang, F.; Nguyen, M.; Qin, F.X.; Tong, Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 2007, 6, 505–514. [Google Scholar] [CrossRef]
- Seawright, J.W.; Samman, Y.; Sridharan, V.; Mao, X.W.; Cao, M.; Singh, P.; Melnyk, S.; Koturbash, I.; Nelson, G.A.; Hauer-Jensen, M.; et al. Effects of low-dose rate gamma-irradiation combined with simulated microgravity on markers of oxidative stress, DNA methylation potential, and remodeling in the mouse heart. PLoS ONE 2017, 12, e0180594. [Google Scholar] [CrossRef] [PubMed]
- Ricklin, D.; Lambris, J.D. Complement in immune and inflammatory disorders: Pathophysiological mechanisms. J. Immunol. 2013, 190, 3831–3838. [Google Scholar] [CrossRef]
- Bjerre, M.; Hansen, T.K.; Flyvbjerg, A. Complement activation and cardiovascular disease. Horm. Metab. Res. 2008, 40, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.I.; Skjoedt, M.O.; Siow Tan, Y.; Rosbjerg, A.; Garred, P.; Stahl, G.L. Endogenous and natural complement inhibitor attenuates myocardial injury and arterial thrombogenesis. Circulation 2012, 126, 2227–2235. [Google Scholar] [CrossRef]
- Sekine, H.; Kinser, T.T.; Qiao, F.; Martinez, E.; Paulling, E.; Ruiz, P.; Gilkeson, G.S.; Tomlinson, S. The benefit of targeted and selective inhibition of the alternative complement pathway for modulating autoimmunity and renal disease in MRL/lpr mice. Arthritis Rheum. 2011, 63, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Qi, X.; Li, Q.; Jia, W.; Wei, L.; Huang, A.; Liu, K.; Li, Z. Increased complements and high-sensitivity C-reactive protein predict heart failure in acute myocardial infarction. Biomed. Rep. 2016, 5, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cai, D. Complement components sC5b-9 and CH50 predict prognosis in heart failure patients combined with hypertension. Am. J. Hypertens. 2019, 33, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.A.L.; Patzelt, J.; Sauter, M.; Maier, P.; Gekeler, S.; Klingel, K.; Kandolf, R.; Seizer, P.; Gawaz, M.; Geisler, T.; et al. Myocardial expression of the anaphylatoxin receptor C3aR is associated with cardiac inflammation and prognosis in patients with non-ischaemic heart failure. ESC Heart Fail. 2018, 5, 846–857. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, V.C.; Timmers, L.; Van Duijvenvoorde, A.; De Jager, S.C.; Van Middelaar, B.J.; Smeets, M.B.; Woodruff, T.M.; Doevendans, P.A.; Pasterkamp, G.; Hack, C.E.; et al. Leucocyte expression of complement C5a receptors exacerbates infarct size after myocardial reperfusion injury. Cardiovasc. Res. 2014, 103, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Herzog, C.; Larmann, J.; Schmitz, M.; Hilfiker-Kleiner, D.; Gessner, J.E.; Theilmeier, G. The receptor for activated complement factor 5 (C5aR) conveys myocardial ischemic damage by mediating neutrophil transmigration. Immunobiology 2013, 218, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Jenei, Z.M.; Zima, E.; Csuka, D.; Munthe-Fog, L.; Hein, E.; Szeplaki, G.; Becker, D.; Karadi, I.; Prohaszka, Z.; Garred, P.; et al. Complement activation and its prognostic role in post-cardiac arrest patients. Scand. J. Immunol. 2014, 79, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Kashirina, D.N.; Percy, A.J.; Pastushkova, L.K.; Borchers, C.H.; Kireev, K.S.; Ivanisenko, V.A.; Kononikhin, A.S.; Nikolaev, E.N.; Larina, I.M. The molecular mechanisms driving physiological changes after long duration space flights revealed by quantitative analysis of human blood proteins. BMC Med. Genom. 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Shi, J.; Zhang, P.; Wang, K.; Li, J.; Liu, H.; Zhou, Y.; Xu, X.; Hao, J.; Sun, X.; et al. Simulated microgravity disrupts intestinal homeostasis and increases colitis susceptibility. FASEB J. 2015, 29, 3263–3273. [Google Scholar] [CrossRef]
- Zhou, Y.; Ni, H.; Li, M.; Sanzari, J.K.; Diffenderfer, E.S.; Lin, L.; Kennedy, A.R.; Weissman, D. Effect of solar particle event radiation and hindlimb suspension on gastrointestinal tract bacterial translocation and immune activation. PLoS ONE 2012, 7, e44329. [Google Scholar] [CrossRef]
- Jin, M.; Zhang, H.; Zhao, K.; Xu, C.; Shao, D.; Huang, Q.; Shi, J.; Yang, H. Responses of Intestinal Mucosal Barrier Functions of Rats to Simulated Weightlessness. Front. Physiol. 2018, 9, 729. [Google Scholar] [CrossRef] [PubMed]
- Knuefermann, P.; Schwederski, M.; Velten, M.; Krings, P.; Ehrentraut, H.; Rudiger, M.; Boehm, O.; Fink, K.; Dreiner, U.; Grohe, C.; et al. Bacterial DNA induces myocardial inflammation and reduces cardiomyocyte contractility: Role of toll-like receptor 9. Cardiovasc. Res. 2008, 78, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.D.; Ramos, R.A.; Tobias, P.S.; Ulevitch, R.J.; Mathison, J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 1990, 249, 1431–1433. [Google Scholar] [CrossRef]
- Guo, L.; Akahori, H.; Harari, E.; Smith, S.L.; Polavarapu, R.; Karmali, V.; Otsuka, F.; Gannon, R.L.; Braumann, R.E.; Dickinson, M.H.; et al. CD163+ macrophages promote angiogenesis and vascular permeability accompanied by inflammation in atherosclerosis. J. Clin. Investig. 2018, 128, 1106–1124. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.A.; Sulciner, M.L.; Nowicki, K.D.; Miller, A.D.; Burdo, T.H.; Williams, K.C. Elevated numbers of CD163+ macrophages in hearts of simian immunodeficiency virus-infected monkeys correlate with cardiac pathology and fibrosis. AIDS Res. Hum. Retroviruses 2014, 30, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Seawright, J.W.; Sridharan, V.; Landes, R.D.; Cao, M.; Singh, P.; Koturbash, I.; Mao, X.W.; Miousse, I.R.; Singh, S.P.; Nelson, G.A.; et al. Effects of low-dose oxygen ions and protons on cardiac function and structure in male C57BL/6J mice. Life Sci. Space Res. 2019, 20, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, L.; Kiffer, F.; Puukila, S.; Lowe, M.G.; Goo, B.; Luthens, A.; Schreurs, A.S.; Torres, S.M.; Steczina, S.; Tahimic, C.G.T.; et al. Mitochondria-Targeted Human Catalase in the Mouse Longevity MCAT Model Mitigates Head-Tilt Bedrest-Induced Neuro-Inflammation in the Hippocampus. Life 2022, 12, 1838. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Blackwell, V.K.; Harandi, B.; Gibbons, A.C.; Siu, O.; Irby, I.; Rees, A.; Cornejal, N.; Sattler, K.M.; Sheng, T.; et al. Influence of the spaceflight environment on macrophage lineages. NPJ Microgravity 2024, 10, 63. [Google Scholar] [CrossRef]
- Yu, L.; Feng, Z. The Role of Toll-Like Receptor Signaling in the Progression of Heart Failure. Mediators Inflamm. 2018, 2018, 9874109. [Google Scholar] [CrossRef]
- Parizadeh, S.M.; Ghandehari, M.; Heydari-Majd, M.; Seifi, S.; Mardani, R.; Parizadeh, S.M.; Ghayour-Mobarhan, M.; Ferns, G.A.; Hassanian, S.M.; Avan, A. Toll-like Receptors Signaling Pathways as a Potential Therapeutic Target in Cardiovascular Disease. Curr. Pharm. Des. 2018, 24, 1887–1898. [Google Scholar] [CrossRef]
- Satoh, M.; Akatsu, T.; Ishikawa, Y.; Minami, Y.; Takahashi, Y.; Nakamura, M. Association between toll-like receptor 8 expression and adverse clinical outcomes in patients with enterovirus-associated dilated cardiomyopathy. Am. Heart J. 2007, 154, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, A.; Das, P.; Hashimoto, I.; Nakao, T.; Deguchi, Y.; Gouraud, S.S.; Waki, H.; Muragaki, Y.; Maeda, M. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS ONE 2014, 9, e112811. [Google Scholar] [CrossRef] [PubMed]
- Bonney, S.; Kominsky, D.; Brodsky, K.; Eltzschig, H.; Walker, L.; Eckle, T. Cardiac Per2 functions as novel link between fatty acid metabolism and myocardial inflammation during ischemia and reperfusion injury of the heart. PLoS ONE 2013, 8, e71493. [Google Scholar] [CrossRef]
- Flynn-Evans, E.E.; Barger, L.K.; Kubey, A.A.; Sullivan, J.P.; Czeisler, C.A. Circadian misalignment affects sleep and medication use before and during spaceflight. NPJ Microgravity 2016, 2, 15019. [Google Scholar] [CrossRef] [PubMed]
- Fujita, S.I.; Rutter, L.; Ong, Q.; Muratani, M. Integrated RNA-seq Analysis Indicates Asynchrony in Clock Genes between Tissues under Spaceflight. Life 2020, 10, 196. [Google Scholar] [CrossRef]
- Ma, Y.; Kong, L.; Nan, K.; Qi, S.; Ru, L.; Ding, C.; Wang, D. Apolipoprotein-J prevents angiotensin II-induced apoptosis in neonatal rat ventricular cells. Lipids Health Dis. 2015, 14, 114. [Google Scholar] [CrossRef]
- Hirschenson, J.; Melgar-Bermudez, E.; Mailloux, R.J. The Uncoupling Proteins: A Systematic Review on the Mechanism Used in the Prevention of Oxidative Stress. Antioxidants 2022, 11, 322. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, G.; Fasciolo, G.; Magnacca, N.; Goglia, F.; Lombardi, A.; Venditti, P. Oxidative damage and mitochondrial functionality in hearts from KO UCP3 mice housed at thermoneutrality. J. Physiol. Biochem. 2022, 78, 415–425. [Google Scholar] [CrossRef]
- da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B.; et al. Comprehensive Multi-omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 2020, 183, 1185–1201. [Google Scholar] [CrossRef]
- Indo, H.P.; Majima, H.J.; Terada, M.; Suenaga, S.; Tomita, K.; Yamada, S.; Higashibata, A.; Ishioka, N.; Kanekura, T.; Nonaka, I.; et al. Changes in mitochondrial homeostasis and redox status in astronauts following long stays in space. Sci. Rep. 2016, 6, 39015. [Google Scholar] [CrossRef]
- Zhuang, J.; Chen, L.; Li, G.; Xia, L.; Wu, S.; Leng, J.; Tao, X.; Hong, J.; Wu, Y.; Wang, S.; et al. RCAN1 deficiency aggravates sepsis-induced cardiac remodeling and dysfunction by accelerating mitochondrial pathological fission. Inflamm. Res. 2022, 71, 1589–1602. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Y.; Qiu, K.; Zhu, J.; Wu, Y. RCAN1 in cardiovascular diseases: Molecular mechanisms and a potential therapeutic target. Mol. Med. 2020, 26, 118. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Tahimic, C.G.T.; Almeida, E.A.C.; Globus, R.K. Spaceflight Modulates the Expression of Key Oxidative Stress and Cell Cycle Related Genes in Heart. Int. J. Mol. Sci. 2021, 22, 9088. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).