Comorbidity-Guided Text Mining and Omics Pipeline to Identify Candidate Genes and Drugs for Alzheimer’s Disease
Abstract
1. Introduction
- (i)
- The proposed approach integrates text mining and genomics together to find common genes and drugs for AD and its comorbid diseases. The existing works either use text mining or genomics to find new treatment options for a disease. The existing works do not consider the comorbid diseases too. Thus, we advance the existing research methods for finding new treatment options.
- (ii)
- The proposed approach queries PubMed abstracts to extract comorbid diseases for AD. The existing approaches extract the comorbid diseases for a disease of interest from patients’ electronic health records (EHRs). However, accessing EHRs requires institutional revenue board (IRB) approval. There are other challenges too. Unlike patients’ EHRs, PubMed is an open source and it is free to use.
- (iii)
- A comorbidity-guided approach is proposed to identify new candidate genes and drugs for AD and its comorbid diseases. To our knowledge, the existing works on identifying new candidate genes and drugs for a disease, especially AD, do not consider its comorbid diseases. However, the knowledge on comorbid diseases, their candidate genes, and drugs prescribed for treating these comorbid diseases is important to avoid drug–drug interaction.
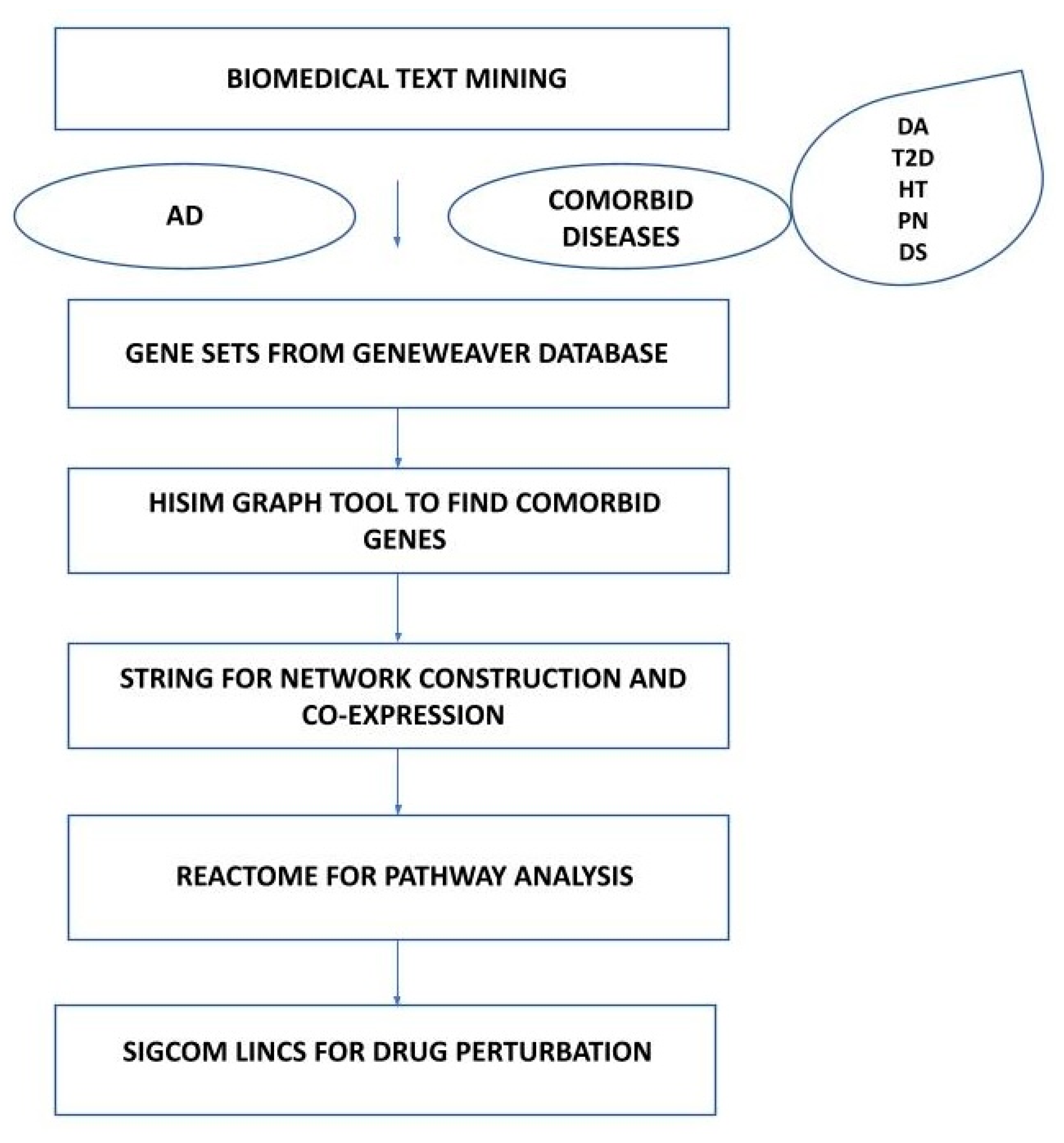
2. Materials and Methods
2.1. Text Mining Approach
2.1.1. Lexicon of MeSH Diseases
2.1.2. Retrieving Comorbid Diseases from PubMed
- (i)
- Number of PubMed articles with AD and a co-occurring disease in the MeSH index;
- (ii)
- Number of PubMed articles with the co-occurring disease in the MeSH index;
- (iii)
- Number of PubMed articles with AD in the MeSH index;
- (iv)
- Total number of PubMed articles (125,924, access date 22 November 2022) with the MeSH term “Comorbidity” in the MeSH index.
- Boolean query (a) for count (i): “Alzheimer disease” [MeSH] AND “Comorbidity” [MeSH] AND “CO-OCCURRING DISEASE” [MeSH];
- Boolean query (b) for count (ii): “CO-OCCURRING DISEASE” [MeSH] AND “Comorbidity” [MeSH] NOT “Alzheimer disease” [MeSH];
- Boolean query (c) for count (iii): “Alzheimer disease” [MeSH] AND “Comorbidity” [MeSH] NOT “CO-OCCURRING DISEASE” [MeSH];
- Boolean query (d) for count (iv): “Comorbidity” [MeSH].
2.2. Genomics Approach
2.2.1. Identification of Common Genes for AD and Five Comorbid Diseases
2.2.2. Protein–Protein Interaction from STRING Database
2.2.3. Enrichment Analysis for Pathways
2.2.4. Drugs from Library of Integrated Network-Based Cellular Signatures (Sigcom LINCS)
3. Results
3.1. Comorbid Diseases for AD
3.2. Common Genes among AD and Comorbid Diseases
3.3. Network Analysis of Common Genes
3.4. Comorbid Diseases and Drug Perturbation
4. Discussion
4.1. Comorbid Diseases and Genetic Analysis
4.2. Comorbid Diseases and Pathway Analysis
4.3. Comorbid Diseases and Drug Perturbation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bellenguez, C.; Fahri, K.; Iris, E.J.; Luca, K.; Sonia, M.G.; Najaf, A.; Adam, C.N. New Insights into the Genetic Etiology of Alzheimer’s Disease and Related Dementias. Nat. Genet. 2022, 54, 412–436. [Google Scholar] [CrossRef] [PubMed]
- Ridge, P.G.; Mukherjee, S.; Crane, P.K.; Kauwe, J.S.K. Alzheimer’s Disease: Analyzing the Missing Heritability. PLoS ONE 2013, 8, e79771. [Google Scholar] [CrossRef] [PubMed]
- Anand, D.; Manoharan, S.; Iyyappan, O.R.; Anand, S.; Raja, K. Extracting Significant Comorbid Diseases from MeSH Index of PubMed. Methods Mol. Biol. 2022, 2496, 283–299. [Google Scholar] [CrossRef]
- Berman, A.N.; Biery, D.W.; Ginder, C.; Hulme, O.L.; Marcusa, D.; Leiva, O.; Wu, W.Y. Natural language processing for the assessment of cardiovascular disease comorbidities: The cardio-Canary comorbidity project. Clin. Cardiol. 2021, 44, 1296–1304. [Google Scholar] [CrossRef]
- Al-Harrasi, A.M.; Iqbal, E.; Tsamakis, K.; Lasek, J.; Gadelrab, R.; Soysal, P.; Kohlhoff, E. Motor signs in Alzheimer’s disease and vascular dementia: Detection through natural language processing, comorbid features and relationship to adverse outcomes. Exp. Gerontol. 2021, 146, 111223. [Google Scholar] [CrossRef]
- Anand, S.; Iyyappan, O.R.; Manoharan, S.; Anand, D.; Jose, M.A.; Shanker, R.R. Text Mining Protocol to Retrieve Significant Drug–Gene Interactions from PubMed Abstracts. Methods Mol. Biol. 2022, 2496, 17–39. [Google Scholar] [CrossRef] [PubMed]
- Fertig, E.J.; Slebos, R.; Chung, C.H. Application of Genomic and Proteomic Technologies in Biomarker Discovery. Am. Soc. Clin. Oncol. Educ. 2012, 32, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Keenan, A.B.; Wojciechowicz, M.L.; Wang, Z.; Jagodnik, K.M.; Jenkins, S.L.; Lachmann, A.; Avi, M. Connectivity Mapping: Methods and Applications. Annu. Rev. Biomed. Data Sci. 2019, 2, 69–92. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Prabahar, A.; Palanisamy, A. A Hybrid Protocol for Identifying Comorbidity-Based Potential Drugs for COVID-19 Using Biomedical Literature Mining, Network Analysis, and Deep Learning. Methods Mol. Biol. 2022, 2496, 203–219. [Google Scholar] [CrossRef]
- Adams, C.M.; Ebert, S.M.; Dyle, M.C. Use of mRNA expression signatures to discover small molecule inhibitors of skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 263–268. [Google Scholar] [CrossRef]
- Baker, E.J.; Jay, J.J.; Bubier, J.A.; Langston, M.A.; Chesler, E.J. GeneWeaver: A web-based system for integrative functional genomics. Nucleic Acids Res. 2012, 40, D1067–D1076. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, J.E. SigCom LINCS: Data and metadata search engine for a million gene expression signatures. Nucleic Acids Res. 2022, 50, W697–W709. [Google Scholar] [CrossRef] [PubMed]
- Barber, R.C. The Genetics of Alzheimer’s Disease. Scientifica 2012, 2012, 246210. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenomics of Cognitive Dysfunction and Neuropsychiatric Disorders in Dementia. Int. J. Mol. Sci. 2020, 21, 3059. [Google Scholar] [CrossRef] [PubMed]
- Seto, M.; Weiner, R.L.; Dumitrescu, L.; Hohman, T.J. Protective genes and pathways in Alzheimer’s disease: Moving towards precision interventions. Mol. Neurodegener. 2021, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.J.; Kim, M.-J.; Kim, J.; Ryu, H.-S.; Kim, Y.J.; Kim, S.Y.; Lee, J.-H. Association of type 2 diabetes GWAS loci and the risk of Parkinson’s and Alzheimer’s diseases. Parkinsonism Relat. Disord. 2015, 21, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Silzer, T.K.; Phillips, N.R. Etiology of type 2 diabetes and Alzheimer’s disease: Exploring the mitochondria. Mitochondrion 2018, 43, 16–24. [Google Scholar] [CrossRef]
- Maple-Grødem, J.; Chung, J.; Lunde, K.A.; Tzoulis, C.; Tysnes, O.B.; Pedersen, K.F.; Alves, G. Alzheimer disease associated variants in SORL1 accelerate dementia development in Parkinson disease. Neurosci. Lett. 2018, 674, 123–126. [Google Scholar] [CrossRef]
- Prokopenko, I.; Miyakawa, G.; Zheng, B.; Heikkinen, J.; Quayle, D.P.; Udeh-Momoh, C.; Claringbould, A. Alzheimer’s disease pathology explains association between dementia with Lewy bodies and APOE-ε4/TOMM40 long poly-T repeat allele variants. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2019, 5, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Wang, K.; Hu, G.; Wang, X.; Miao, Z.; Azevedo, J.A.; Suh, E. APOE and TREM2 regulate amyloid-responsive microglia in Alzheimer’s disease. Acta Neuropathol. 2020, 140, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Xu, C.; Guo, Q.; Zhu, F.Q. SORL1 mutations are associated with parkinsonian and psychiatric features in Alzheimer disease. Medicine 2021, 100, e25585. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Mao, C.; Liu, C.; Li, J.; Huang, X.; Wang, J.; Lei, D. Association between Common Variants of APOE, ABCA7, A2M, BACE1, and Cerebrospinal Fluid Biomarkers in Alzheimer’s Disease: Data from the PUMCH Dementia Cohort. J. Alzheimer’s Dis. 2022, 85, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- van der Kant, R.; Goldstein, L.S.B. Cellular Functions of the Amyloid Precursor Protein from Development to Dementia. Dev. Cell 2015, 32, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Vance, E.; Gonzalez, J.; Miller, J.B. Failure to detect synergy between variants in transferrin and hemochromatosis and Alzheimer’s disease in large cohort. Neurobiol. Aging 2020, 89, 142.e9–142.e12. [Google Scholar] [CrossRef] [PubMed]
- Ali-Rahmani, F.; Schengrund, C.-L.; Connor, J.R. HFE gene variants, iron, and lipids: A novel connection in Alzheimerâ€TMs disease. Front. Pharmacol. 2014, 5, 165. [Google Scholar] [CrossRef]
- Fortea, J.; Quiroz, Y.T.; Ryan, N.S. Lessons from Down syndrome and autosomal dominant Alzheimer’s disease. Lancet Neurol. 2023, 22, 5–6. [Google Scholar] [CrossRef]
- Iulita, M.F.; Chavez, D.G.; Christensen, M.K.; Tamayo, N.V.; Plana-Ripoll, O.; Rasmussen, S.A.; Figuls, M.R. Association of Alzheimer Disease with Life Expectancy in People with Down Syndrome. JAMA Netw. Open 2022, 5, e2212910. [Google Scholar] [CrossRef]
- Feng, Q.; Luo, Y.; Zhang, X.-N.; Yang, X.-F.; Hong, X.-Y.; Sun, D.-S.; Li, X.-C. MAPT/Tau accumulation represses autophagy flux by disrupting IST1-regulated ESCRT-III complex formation: A vicious cycle in Alzheimer neurodegeneration. Autophagy 2020, 16, 641–658. [Google Scholar] [CrossRef] [PubMed]
- Cuyvers, E.; Sleegers, K. Genetic variations underlying Alzheimer’s disease: Evidence from genome-wide association studies and beyond. Lancet Neurol. 2016, 15, 857–868. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B.; De Strooper, B.; Annaert, W.; Cupers, P.; Saftig, P.; Craessaerts, K.; Mumm, J.S. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 1999, 398, 518–522. [Google Scholar] [CrossRef]
- Kapoor, A.; Nation, D.A. Role of Notch signaling in neurovascular aging and Alzheimer’s disease. Semin. Cell Dev. Biol. 2021, 116, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Babicheva, A.; Yuan, J.X.-J. Endothelial Notch1 in Pulmonary Hypertension. Circ. Res. 2019, 124, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ou, R.; Chen, Y.; Gu, X.; Wei, Q.; Cao, B.; Zhang, L. Mutation analysis of TMEM family members for early-onset Parkinson’s disease in Chinese population. Neurobiol. Aging 2021, 101, 299.e1–299.e6. [Google Scholar] [CrossRef]
- Barthelson, K.; Newman, M.; Lardelli, M. Sorting Out the Role of the Sortilin-Related Receptor 1 in Alzheimer’s Disease. J. Alzheimers Dis. Rep. 2020, 4, 123–140. [Google Scholar] [CrossRef] [PubMed]
- de la Monte, S.M.; Chiche, J.-D.; von dem Bussche, A.; Sanyal, S.; Lahousse, S.A.; Janssens, S.P.; Bloch, K.D. Nitric Oxide Synthase-3 Overexpression Causes Apoptosis and Impairs Neuronal Mitochondrial Function: Relevance to Alzheimer’s-Type Neurodegeneration. Lab. Investig. 2003, 83, 287–298. [Google Scholar] [CrossRef][Green Version]
- Singleton, A.; Gibson, A.M.; McKeith, I.G.; Ballard, C.G.; Edwardson, J.A.; Morris, C.M. Nitric oxide synthase gene polymorphisms in Alzheimer’s disease and dementia with Lewy bodies. Neurosci. Lett. 2001, 303, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.C.; Choe, Y.M.; Suh, G.-H.; Choi, I.-G.; Lee, J.H.; Kim, H.S.; Hwang, J.; Yi, D.; Kim, J.W. A combination of midlife diabetes mellitus and the apolipoprotein E ε4 allele increase risk for cognitive decline. Front. Aging Neurosci. 2022, 14, 1065117. [Google Scholar] [CrossRef]
- Takechi, R.; Sharif, A.; Brook, E.; Majimbi, M.; Chan, D.C.; Lam, V.; Watts, G.F.; Mamo, J.C.L. Is type 2 diabetes associated dementia a microvascular early-Alzheimer’s phenotype induced by aberrations in the peripheral metabolism of lipoprotein-amyloid? Front. Endocrinol. 2023, 14, 1127481. [Google Scholar] [CrossRef]

| Co-Occurring Disease | PubMed Articles with AD and Co-Occurring Disease (P) | PubMed Articles with AD and All Co-Occurring Diseases (T) | Sort Ratio (P/T) |
|---|---|---|---|
| Dementia | 142 | 843 | 0.1684 |
| Type 2 diabetes | 95 | 843 | 0.1127 |
| Hypertension | 60 | 843 | 0.0712 |
| Parkinson’s disease | 51 | 843 | 0.0605 |
| Down Syndrome | 43 | 843 | 0.0510 |
| Comorbid Disease (CD) | PubMed Articles | FET p-Value | Literature Evidence (PMID) | |||
|---|---|---|---|---|---|---|
| AD and CD | CD | AD | Comorbidity | |||
| Dementia | 142 | 220 | 1066 | 125,924 | 1.30 × 10−238 | 11406927 |
| Type 2 diabetes | 95 | 11,209 | 1113 | 125,924 | 0.68 | 28922161 |
| Hypertension | 60 | 6726 | 1148 | 125,924 | 0.59 | 33888050 |
| Parkinson’s disease | 51 | 683 | 1157 | 125,924 | 6.28 × 10−30 | 26820182 |
| Down syndrome | 43 | 346 | 1165 | 125,924 | 1.88 × 10−34 | 28009725 |
| Multimorbid Diseases | Number of Common Genes | Common Gene(s) |
|---|---|---|
| AD, dementia, Parkinson’s disease, Down syndrome | 4 | MAPT, PSEN1, APP, APOE |
| AD, dementia, Parkinson’s disease | 5 | A2M, ABCA7, PLAU, PSEN2, MPO |
| AD, dementia, Down syndrome | 2 | COL18A1, SYNJ1 |
| AD, dementia, hypertension, Parkinson’s disease | 2 | HFE, NOS3 |
| AD, Parkinson’s disease, Down syndrome | 1 | SORL1 |
| AD, type 2 diabetes, hypertension | 1 | NOTCH2 |
| AD, dementia, hypertension | 1 | TGFB2 |
| AD, dementia, type 2 diabetes | 1 | LAMA1 |
| Comorbid Diseases | Common Genes | Pathways for Common Genes (Reactome) (p < 0.05) | PPI Enrichment (STRING) (p-Value) |
|---|---|---|---|
| AD, dementia | 41 | 28 | <1.0 × 10−16 |
| AD, type 2 diabetes | 5 | 36 | 3.56 × 10−5 |
| AD, hypertension | 26 | 33 | 3.33 × 10−16 |
| AD, Parkinson’s disease | 22 | 27 | <1.0 × 10−16 |
| AD, Down syndrome | 33 | 92 | <1.0 × 10−16 |
| AD and Comorbid Disease | Drug (Top 10) | p-Value |
|---|---|---|
| AD and Dementia | axitinib | 2.19 × 10−7 |
| lamotrigine | 8.11 × 10−7 | |
| ataluren | 1.99 × 10−6 | |
| vandetanib | 2.21 × 10−6 | |
| etofylline | 2.49 × 10−6 | |
| BRD-K18100239 | 3.92 × 10−6 | |
| AS-605240 | 4.53 × 10−6 | |
| velnacrine | 5.96 × 10−6 | |
| BRD-K30758067 | 6.15 × 10−6 | |
| quiflapon | 7.10 × 10−6 | |
| AD and Type 2 Diabetes | BMS-777607 | 0.0021 |
| trifluoperazine | 0.0022 | |
| BRD-K18972207 | 0.0025 | |
| pirenperone | 0.0025 | |
| lonidamine | 0.0026 | |
| ARRY-334543 | 0.0026 | |
| BRD-K84094241 | 0.0027 | |
| BRD-K89952884 | 0.0028 | |
| CYT-997 | 0.0029 | |
| roflumilast | 0.0030 | |
| AD and Hypertension | vernakalant | 5.73 × 10−7 |
| BRD-K40853697 | 1.38 × 10−6 | |
| marimastat | 1.64 × 10−6 | |
| LXR-623 | 2.09 × 10−6 | |
| regorafenib | 2.54 × 10−6 | |
| aclidinium | 6.77 × 10−6 | |
| α-estradiol | 7.31 × 10−6 | |
| BRD-K38373457 | 7.37 × 10−6 | |
| pidotimod | 7.95 × 10−6 | |
| ifenprodil | 8.46 × 10−6 | |
| AD and Parkinson’s disease | bortezomib | 1.30 × 10−6 |
| maprotiline | 2.52 × 10−5 | |
| flupirtine | 8.11 × 10−5 | |
| PD-0325901 | 8.70 × 10−5 | |
| salmeterol | 9.98 × 10−5 | |
| quizartinib | 0.0001 | |
| dapagliflozin | 0.0001 | |
| gefitinib | 0.0001 | |
| lomitapide | 0.0002 | |
| SB-216763 | 0.0002 | |
| AD and Down syndrome | BRD-A01960364 | 3.77 × 10−8 |
| MG-132 | 3.20 × 10−7 | |
| BRD-A06909528 | 6.13 × 10−7 | |
| teniposide | 1.24 × 10−6 | |
| MD-049 | 1.64 × 10−6 | |
| BRD-A95820578 | 1.89 × 10−6 | |
| pravastatin | 5.51 × 10−6 | |
| gatifloxacin | 5.74 × 10−6 | |
| VU-0418942-1 | 6.26 × 10−6 | |
| marimastat | 6.74 × 10−6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oviya, I.R.; Sankar, D.; Manoharan, S.; Prabahar, A.; Raja, K. Comorbidity-Guided Text Mining and Omics Pipeline to Identify Candidate Genes and Drugs for Alzheimer’s Disease. Genes 2024, 15, 614. https://doi.org/10.3390/genes15050614
Oviya IR, Sankar D, Manoharan S, Prabahar A, Raja K. Comorbidity-Guided Text Mining and Omics Pipeline to Identify Candidate Genes and Drugs for Alzheimer’s Disease. Genes. 2024; 15(5):614. https://doi.org/10.3390/genes15050614
Chicago/Turabian StyleOviya, Iyappan Ramalakshmi, Divya Sankar, Sharanya Manoharan, Archana Prabahar, and Kalpana Raja. 2024. "Comorbidity-Guided Text Mining and Omics Pipeline to Identify Candidate Genes and Drugs for Alzheimer’s Disease" Genes 15, no. 5: 614. https://doi.org/10.3390/genes15050614
APA StyleOviya, I. R., Sankar, D., Manoharan, S., Prabahar, A., & Raja, K. (2024). Comorbidity-Guided Text Mining and Omics Pipeline to Identify Candidate Genes and Drugs for Alzheimer’s Disease. Genes, 15(5), 614. https://doi.org/10.3390/genes15050614





