Decoding Evolution of Rubioideae: Plastomes Reveal Sweet Secrets of Codon Usage, Diagnostides, and Superbarcoding
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. Library Preparation and Assembly
2.3. Chloroplast Genome Visualisation, Phylogenetic Analysis, Nucleotide Diversity, and Species Delimitation
2.4. Relative Synonymous Codon Usage
3. Results
3.1. Characteristics of Galium Chloroplast Genomes
3.2. Intraspecific Variation of Plastid Genomes of Galium, Leptodermis, and Rubia
3.3. Molecular Delimitation of Galium, Leptodermis, and Rubia
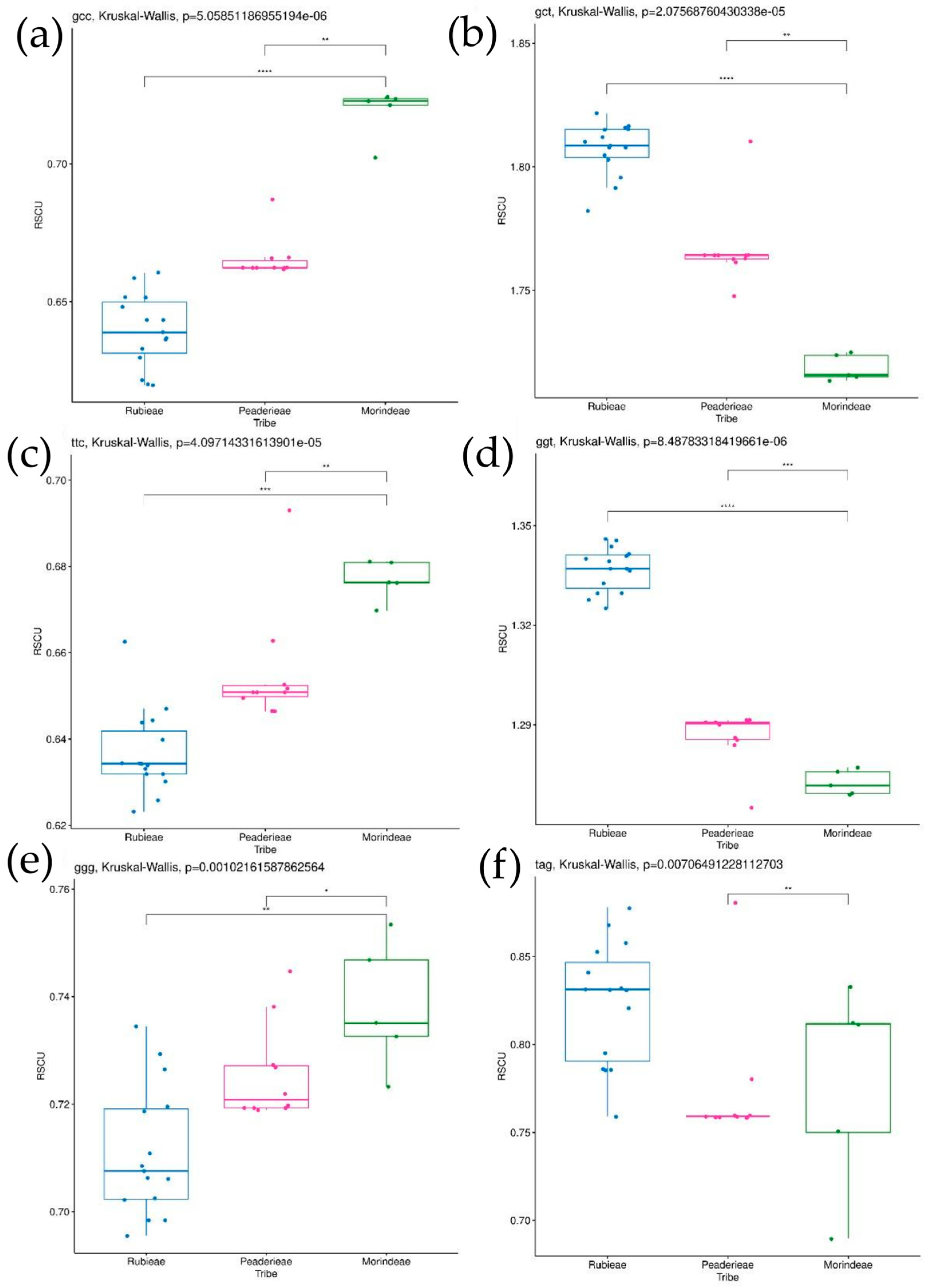
3.4. Codon Usage
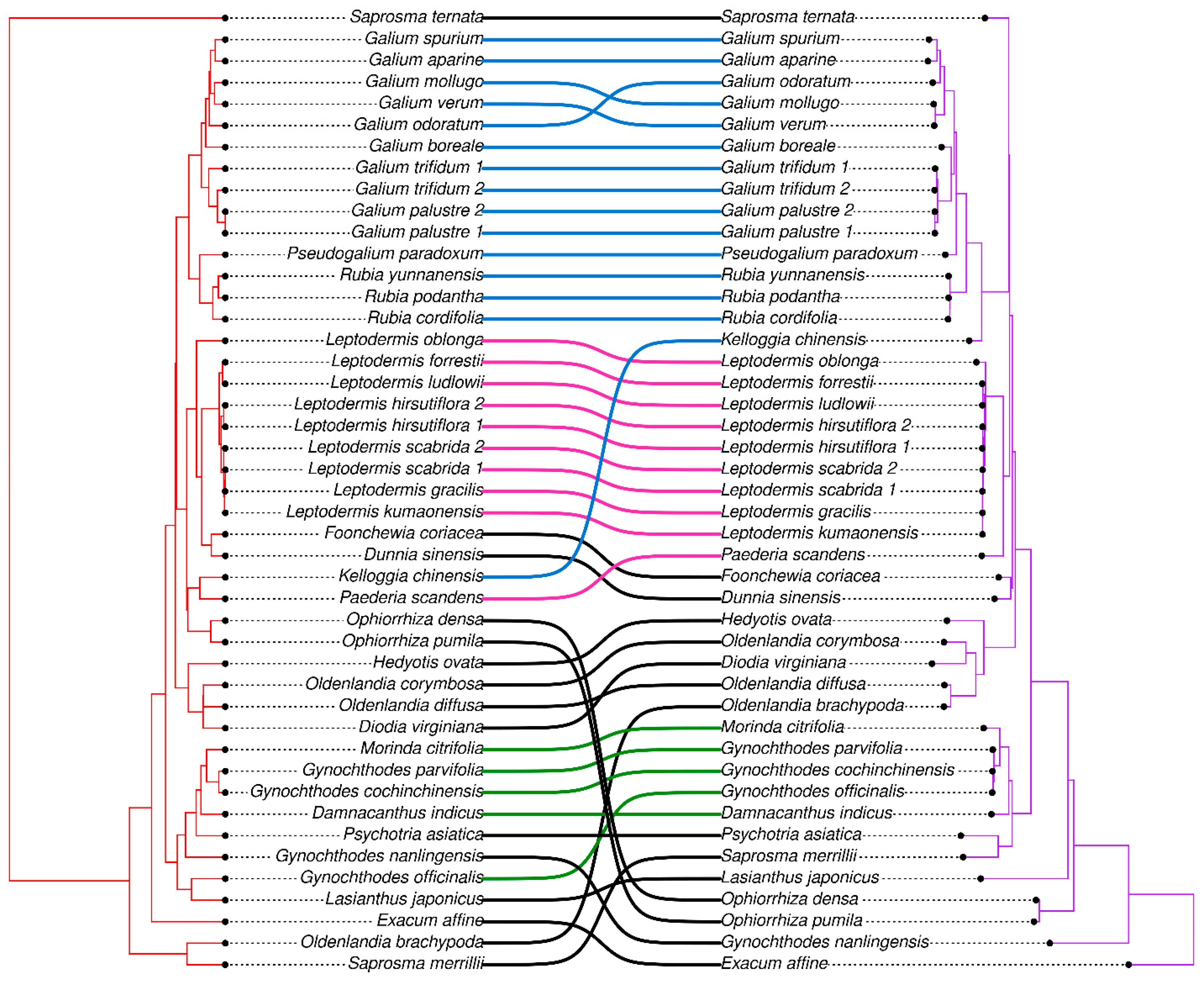
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, A.P.; Govaerts, R.; Bridson, D.M.; Ruhsam, M.; Moat, J.; Brummitt, N.A. A Global Assessment of Distribution, Diversity, Endemism, and Taxonomic Effort in the Rubiaceae1. Ann. Mo. Bot. Gard. 2009, 96, 68–78. [Google Scholar] [CrossRef]
- Govaerts, R. World Checklist of Rubiaceae; Royal Botanic Gardens: London, UK, 2009. [Google Scholar]
- Rutishauser, R. Polymerous Leaf Whorls in Vascular Plants: Developmental Morphology and Fuzziness of Organ Identities. Int. J. Plant Sci. 1999, 160, S81–S103. [Google Scholar] [CrossRef] [PubMed]
- Preston, C.D. Book Review: Aquatic and Wetland Plants of India, Christopher, D.K. Cook. Oxford University Press, Oxford, 1996. 385pp. Price: £95.00. ISBN 0 19 854821 4. Aquat. Conserv. Mar. Freshw. Ecosyst. 1997, 7, 82–83. [Google Scholar] [CrossRef]
- Martins, D.; Nunez, C.V. Secondary Metabolites from Rubiaceae Species. Molecules 2015, 20, 13422–13495. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Müller, K.F.; Quandt, D. The Evolution of the Plastid Chromosome in Land Plants: Gene Content, Gene Order, Gene Function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Amenu, S.G.; Wei, N.; Wu, L.; Oyebanji, O.; Hu, G.; Zhou, Y.; Wang, Q. Phylogenomic and Comparative Analyses of Coffeeae Alliance (Rubiaceae): Deep Insights into Phylogenetic Relationships and Plastome Evolution. BMC Plant Biol. 2022, 22, 88. [Google Scholar] [CrossRef] [PubMed]
- Ehrendorfer, F.; Barfuss, M.H.J.; Manen, J.-F.; Schneeweiss, G.M. Phylogeny, Character Evolution and Spatiotemporal Diversification of the Species-Rich and World-Wide Distributed Tribe Rubieae (Rubiaceae). PLoS ONE 2018, 13, e0207615. [Google Scholar] [CrossRef]
- Natali, A.; Ehrendorfer, F.; Manen, J.-F. Phylogeny of Rubiaceae-Rubieae Inferred from the Sequence of a cpDNA Intergene Region. Plant Syst. Evol. 1994, 190, 195–211. [Google Scholar] [CrossRef]
- Sawicki, J.; Krawczyk, K.; Kurzyński, M.; Maździarz, M.; Paukszto, Ł.; Sulima, P.; Szczecińska, M. Nanopore Sequencing of Organellar Genomes Revealed Heteroplasmy in Simple Thalloid and Leafy Liverworts. Acta Soc. Bot. Pol. 2023, 92. [Google Scholar] [CrossRef]
- Ciborowski, K.; Skierkowski, B.; Żukowska, K.; Krawczyk, K.; Sawicki, J. Nanopore Sequencing of Chloroplast Genome of Scapania Undulata (L.) Dumort., 1835 (Scapaniaceae, Jungermanniales). Mitochondrial DNA Part B 2022, 7, 1424–1426. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wu, X.; Zhang, D. Comparison of the Abilities of Universal, Super, and Specific DNA Barcodes to Discriminate among the Original Species of Fritillariae Cirrhosae Bulbus and Its Adulterants. PLoS ONE 2020, 15, e0229181. [Google Scholar] [CrossRef] [PubMed]
- Ślipiko, M.; Myszczyński, K.; Buczkowska, K.; Bączkiewicz, A.; Szczecińska, M.; Sawicki, J. Molecular Delimitation of European Leafy Liverworts of the Genus Calypogeia Based on Plastid Super-Barcodes. BMC Plant Biology 2020, 20, 243. [Google Scholar] [CrossRef] [PubMed]
- Teske, D.; Peters, A.; Möllers, A.; Fischer, M. Genomic Profiling: The Strengths and Limitations of Chloroplast Genome-Based Plant Variety Authentication. J. Agric. Food Chem. 2020, 68, 14323–14333. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczakowa, R.; Zarzycki, K.; Mirek, Z.; Pisarek, W. Galium trifidum L. Przytulia trójdzielna: Polish Red Data Book of Plants: Pteridophytes and Flowering Plants; Institute of Nature Conservation of the Polish Academy of Sciences: Kraków, Poland, 2014; pp. 404–406. [Google Scholar]
- Szczecińska, M.; Kwaśniewski, M.; Sawicki, J.; Chwiałkowska, K.; Szandar, K.; Pisarek, W. Development of Microsatellite Markers Using Pyrosequencing in Galium Trifidum (Rubiaceae), a Rare Species in Central Europe. Int. J. Mol. Sci. 2012, 13, 9893–9899. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novoassembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. Organellar Genome DRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Díez Menéndez, C.; Poczai, P.; Williams, B.; Myllys, L.; Amiryousefi, A. IRplus: An Augmented Tool to Detect Inverted Repeats in Plastid Genomes. Genome Biol. Evol. 2023, 15, evad177. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Lo, C.-C.; Li, P.-E.; Davenport, K.W.; Chain, P.S.G. From Raw Reads to Trees: Whole Genome SNP Phylogenetics across the Tree of Life. bioRxiv 2015. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated Protein Sequence and Structural Alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Phylogenetic Tree Visualization. In Data Integration, Manipulation and Visualization of Phylogenetic Trees; Chapman and Hall/CRC: Boca Raton, FL, USA, 2022; pp. 73–96. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating Divergence Times in Large Molecular Phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Tao, Q.; Kumar, S. Theoretical Foundation of the RelTime Method for Estimating Divergence Times from Variable Evolutionary Rates. Mol. Biol. Evol. 2018, 35, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Bremer, B.; Eriksson, T. Time Tree of Rubiaceae: Phylogeny and Dating the Family, Subfamilies, and Tribes. Int. J. Plant Sci. 2009, 170, 766–793. [Google Scholar] [CrossRef]
- Bell, M.A.; Lloyd, G.T. Strap: An R Package for Plotting Phylogenies against Stratigraphy and Assessing Their Stratigraphic Congruence. Palaeontology 2014, 58, 379–389. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.J.; Collins, R.A.; Boyer, S.; Lefort, M.; Malumbres-olarte, J.; Vink, C.J.; Cruickshank, R.H. Spider: An R Package for the Analysis of Species Identity and Evolution, with Particular Reference to DNA Barcoding. Mol. Ecol. Resour. 2012, 12, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, B.; Wittelsbürger, U.; Ramos-Onsins, S.E.; Lercher, M.J. PopGenome: An Efficient Swiss Army Knife for Population Genomic Analyses in R. Mol. Biol. Evol. 2014, 31, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. In Structural Approaches to Sequence Evolution; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. [Google Scholar]
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Mower, J.P.; Vickrey, T.L. Structural Diversity Among Plastid Genomes of Land Plants. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2018; pp. 263–292. [Google Scholar]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Gray, J.C.; Morden, C.W.; et al. Many Parallel Losses of infA from Chloroplast DNA during Angiosperm Evolution with Multiple Independent Transfers to the Nucleus. Plant Cell 2001, 13, 645. [Google Scholar] [CrossRef] [PubMed]
- Dann, M.; Bellot, S.; Schepella, S.; Schaefer, H.; Tellier, A. Mutation Rates in Seeds and Seed-Banking Influence Substitution Rates across the Angiosperm Phylogeny. bioRxiv 2017, 156398. [Google Scholar] [CrossRef]
- Yin, H.; Huang, K.; Xie, P.; Mo, P.; Zhang, N.; Wang, Y. Characterization and Phylogenetic Analysis of the Chloroplast Genome of Galium Spurium. Mitochondrial DNA Part B 2023, 8, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-E.; Li, X.-J.; Peng, D.-L. The Complete Plastid Genome of Kelloggia Chinensis Franch. (Rubiaceae), an Endemic Species from East Asia. Mitochondrial DNA Part B 2021, 7, 91–92. [Google Scholar] [CrossRef]
- Zhao, S.; Liang, H.; Tang, P.; Muchuku, J.K. A Complete Chloroplast Genome of Rubia Yunnanensis Diels (Rubiaceae), a Traditional Chinese Herb Endemic to China. Mitochondrial DNA Part B 2022, 7, 1466–1467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.; Guo, Y.; Chen, S.; Xu, X.; Wang, R. Complete Chloroplast Genomes of Leptodermis Scabrida Complex: Comparative Genomic Analyses and Phylogenetic Relationships. Gene 2021, 791, 145715. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.H.; Tran, T.H.; Le, T.D.; Le, T.L.; Hoang, H.; Chu, H.H. The Complete Chloroplast Genome of An Ophiorrhiza Baviensis Drake Species Reveals Its Molecular Structure, Comparative, and Phylogenetic Relationships. Genes 2023, 14, 227. [Google Scholar] [CrossRef] [PubMed]
- Soza, V.L.; Olmstead, R.G. Molecular Systematics of Tribe Rubieae (Rubiaceae): Evolution of Major Clades, Development of Leaf–like Whorls, and Biogeography. Taxon 2010, 59, 755–771. [Google Scholar] [CrossRef]
- Yang, L.-E.; Meng, Y.; Peng, D.-L.; Nie, Z.-L.; Sun, H. Molecular Phylogeny of Galium L. of the Tribe Rubieae (Rubiaceae)—Emphasis on Chinese Species and Recognition of a New Genus Pseudogalium. Mol. Phylogenet. Evol. 2018, 126, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Ball, L.D.; Bedoya, A.M.; Taylor, C.M.; Lagomarsino, L.P. A Target Enrichment Probe Set for Resolving Phylogenetic Relationships in the Coffee Family, Rubiaceae. Appl. Plant Sci. 2023, 11, e11554. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.L.; Forrest, L.L.; Nicholls, J.A.; Kidner, C.A. Retrieval of Hundreds of Nuclear Loci from Herbarium Specimens. Taxon 2016, 65, 1081–1092. [Google Scholar] [CrossRef]
- Ly, S.N.; Garavito, A.; De Block, P.; Asselman, P.; Guyeux, C.; Charr, J.-C.; Janssens, S.; Mouly, A.; Hamon, P.; Guyot, R. Chloroplast Genomes of Rubiaceae: Comparative Genomics and Molecular Phylogeny in Subfamily Ixoroideae. PLoS ONE 2020, 15, e0232295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, T.; Luebert, F.; Xiang, Y.; Huang, C.-H.; Hu, Y.; Rees, M.; Frohlich, M.W.; Qi, J.; Weigend, M.; et al. Asterid Phylogenomics/Phylotranscriptomics Uncover Morphological Evolutionary Histories and Support Phylogenetic Placement for Numerous Whole-Genome Duplications. Mol. Biol. Evol. 2020, 37, 3188–3210. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.-Q.; Zhu, H. Paraphyly and Phylogenetic Relationships in Lasianthus (Rubiaceae) Inferred from Chloroplast Rps16 Data. Bot. Stud. 2007, 48, 227–232. [Google Scholar]
- Oguri, E.; Sugawara, T.; Peng, C.-I.; Yang, T.Y.A.; Murakami, N. Geographical Origin and Sexual-System Evolution of the Androdioecious Plant Gynochthodes Boninensis (Rubiaceae), Endemic to the Bonin Islands, Japan. Mol. Phylogenetics Evol. 2013, 68, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Bautista, M.A.C.; Tao, W.; Zheng, Y.; Deng, Y.; Chen, T.; Miao, S. Chloroplast Genome Organization and Phylogeny of Gynochthodes Cochinchinensis (DC.) Razafim. & B. Bremer (Rubiaceae). Mitochondrial DNA Part B 2021, 6, 261–262. [Google Scholar] [CrossRef]
- Song, Y.-F.; Yang, Q.-H.; Yi, X.-G.; Zhu, Z.-Q.; Wang, X.-R.; Li, M. Comparative Analysis of Codon Usage Patterns in Chloroplast Genomes of Cherries. Forests 2022, 13, 1891. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, B.; Li, B.; Zhou, Q.; Wang, G.; Jiang, X.; Wang, C.; Xu, Z. Comparative Analysis of Codon Usage Patterns in Chloroplast Genomes of Six Euphorbiaceae Species. PeerJ 2020, 8, e8251. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, D.; He, X.; Yang, Y.; Li, X. Comparative Analysis of the Complete Chloroplast Genomes in Allium Section Bromatorrhiza Species (Amaryllidaceae): Phylogenetic Relationship and Adaptive Evolution. Genes 2022, 13, 1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, Y.; Wang, Y.; Chen, Y.; Dong, W.; Zhang, Z. Complete Chloroplast Genome of Hypericum Perforatum and Dynamic Evolution in Hypericum (Hypericaceae). Int. J. Mol. Sci. 2023, 24, 16130. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-K.; Liu, Y.; Zheng, H.-Y.; Tang, M.-Q.; Xie, S.-Q. Comparative Analysis of Codon Usage Patterns in Nuclear and Chloroplast Genome of Dalbergia (Fabaceae). Genes 2023, 14, 1110. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, Q.; Wang, Y.; Li, M.; Wang, C.; Wang, Z.; Jiao, C.; Xu, C.; Wang, H.; Zhang, Z. Comparative Analysis of Codon Bias in the Chloroplast Genomes of Theaceae Species. Front. Genet. 2022, 13, 824610. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Simmons, M.P.; But, P.P.; Shaw, P.; Wang, R. Application of DNA Barcodes in Hedyotis L. (Spermacoceae, Rubiaceae). J. Syst. Evol. 2011, 49, 203–212. [Google Scholar] [CrossRef]
- Barrabé, L.; Buerki, S.; Mouly, A.; Davis, A.P.; Munzinger, J.; Maggia, L. Delimitation of the Genus Margaritopsis (Rubiaceae) in the Asian, Australasian and Pacific Region, Based on Molecular Phylogenetic Inference and Morphology. Taxon 2012, 61, 1251–1268. [Google Scholar] [CrossRef]
- Alejandro, G.J.; Arshed, M.J.; Valdez, M. Evaluating the Feasibility of Five Candidate DNA Barcoding Loci for Philippine Lasianthus Jack (Lasiantheae: Rubiaceae). Pharmacogn. Mag. 2017, 13, 553. [Google Scholar] [CrossRef] [PubMed]
- Panaligan, A.C.; Baltazar, M.D.; Alejandro, G.J.D. Molecular Authentication of Commercially Cultivated Coffee (Coffea spp.) in the Philippines Using DNA Barcodes. Int. J. Agric. Biol. 2021, 25, 227–230. [Google Scholar]
- Ślipiko, M.; Myszczyński, K.; Buczkowska, K.; Bączkiewicz, A.; Sawicki, J. Super-Mitobarcoding in Plant Species Identification? It Can Work! The Case of Leafy Liverworts Belonging to the Genus Calypogeia. Int. J. Mol. Sci. 2022, 23, 15570. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, J.; Krawczyk, K.; Ślipiko, M.; Szandar, K.; Szczecińska, M. Comparative Analysis of Apopellia Endiviifolia Plastomes Reveals a Strikingly High Level of Differentiation between Its Terrestrial and Water Form. Diversity 2021, 13, 674. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the Most Promising Plastid DNA Barcode of Land Plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, S.K.; Delprete, P.G.; Neupane, S.; Motley, T.J. Molecular Phylogenetic Analysis and Generic Delimitations in Tribe Chiococceae (Cinchonoideae, Rubiaceae). Bot. J. Linn. Soc. 2018, 187, 365–396. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciborowski, K.; Szczecińska, M.; Maździarz, M.; Sawicki, J.; Paukszto, Ł. Decoding Evolution of Rubioideae: Plastomes Reveal Sweet Secrets of Codon Usage, Diagnostides, and Superbarcoding. Genes 2024, 15, 562. https://doi.org/10.3390/genes15050562
Ciborowski K, Szczecińska M, Maździarz M, Sawicki J, Paukszto Ł. Decoding Evolution of Rubioideae: Plastomes Reveal Sweet Secrets of Codon Usage, Diagnostides, and Superbarcoding. Genes. 2024; 15(5):562. https://doi.org/10.3390/genes15050562
Chicago/Turabian StyleCiborowski, Kamil, Monika Szczecińska, Mateusz Maździarz, Jakub Sawicki, and Łukasz Paukszto. 2024. "Decoding Evolution of Rubioideae: Plastomes Reveal Sweet Secrets of Codon Usage, Diagnostides, and Superbarcoding" Genes 15, no. 5: 562. https://doi.org/10.3390/genes15050562
APA StyleCiborowski, K., Szczecińska, M., Maździarz, M., Sawicki, J., & Paukszto, Ł. (2024). Decoding Evolution of Rubioideae: Plastomes Reveal Sweet Secrets of Codon Usage, Diagnostides, and Superbarcoding. Genes, 15(5), 562. https://doi.org/10.3390/genes15050562





