Genome-Wide Investigation of the PLD Gene Family in Tomato: Identification, Analysis, and Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment Methods
2.2. Identification of the PLD Gene Family in Tomato
2.3. Phylogenetic Relationships, Gene Structure, Domains, Protein Motifs, and Cis-Regulatory Elements of the SlPLD Gene Family
2.4. Synteny Analysis of SlPLD Genes
2.5. Expression Patterns of SlPLD Genes
2.6. RNA Extraction and Quantitative Real-Time PCR Analyses
3. Results
3.1. Identification and Analysis of SlPLD Family Members in Tomato
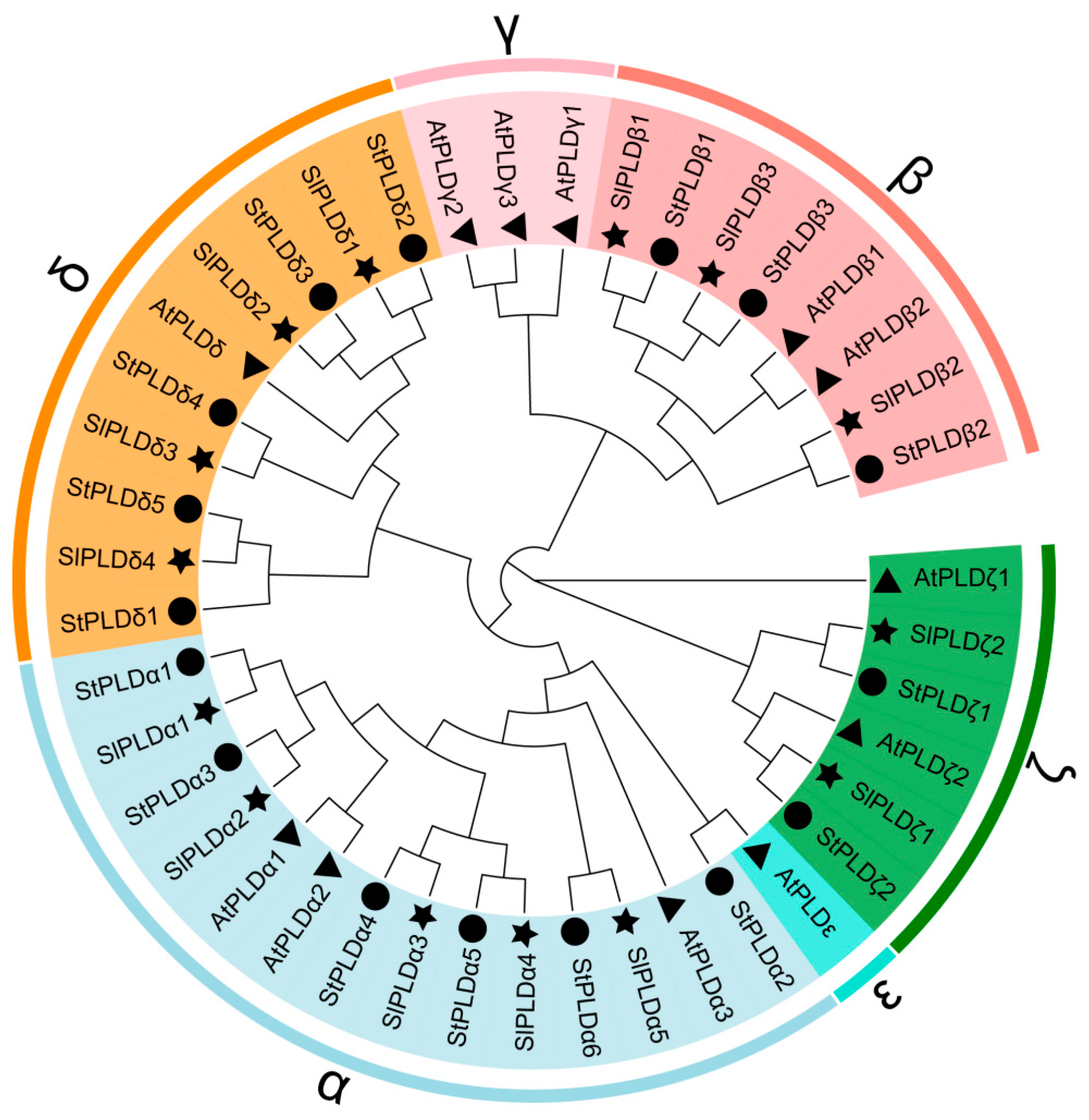
3.2. Phylogenetic Analysis of SlPLD Proteins
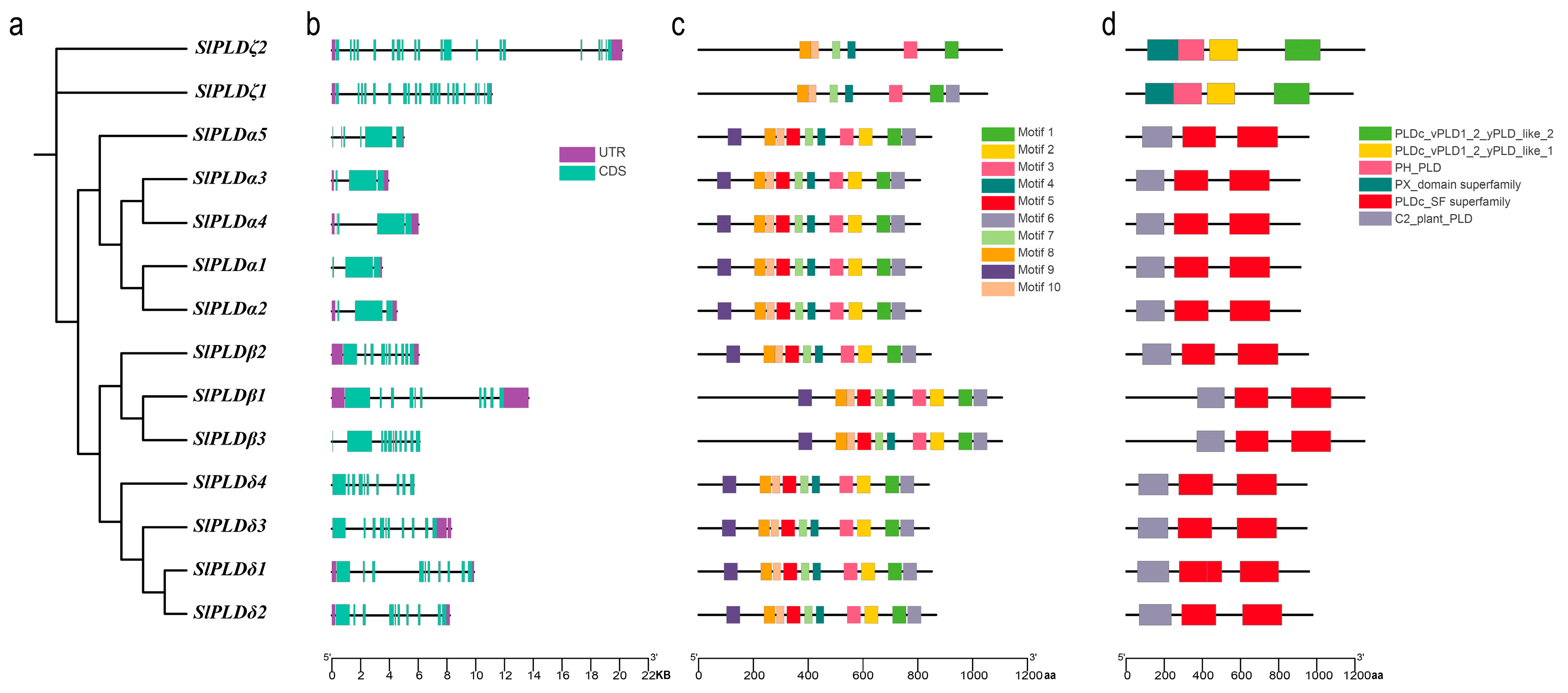
3.3. Gene and Domain Structure Analysis
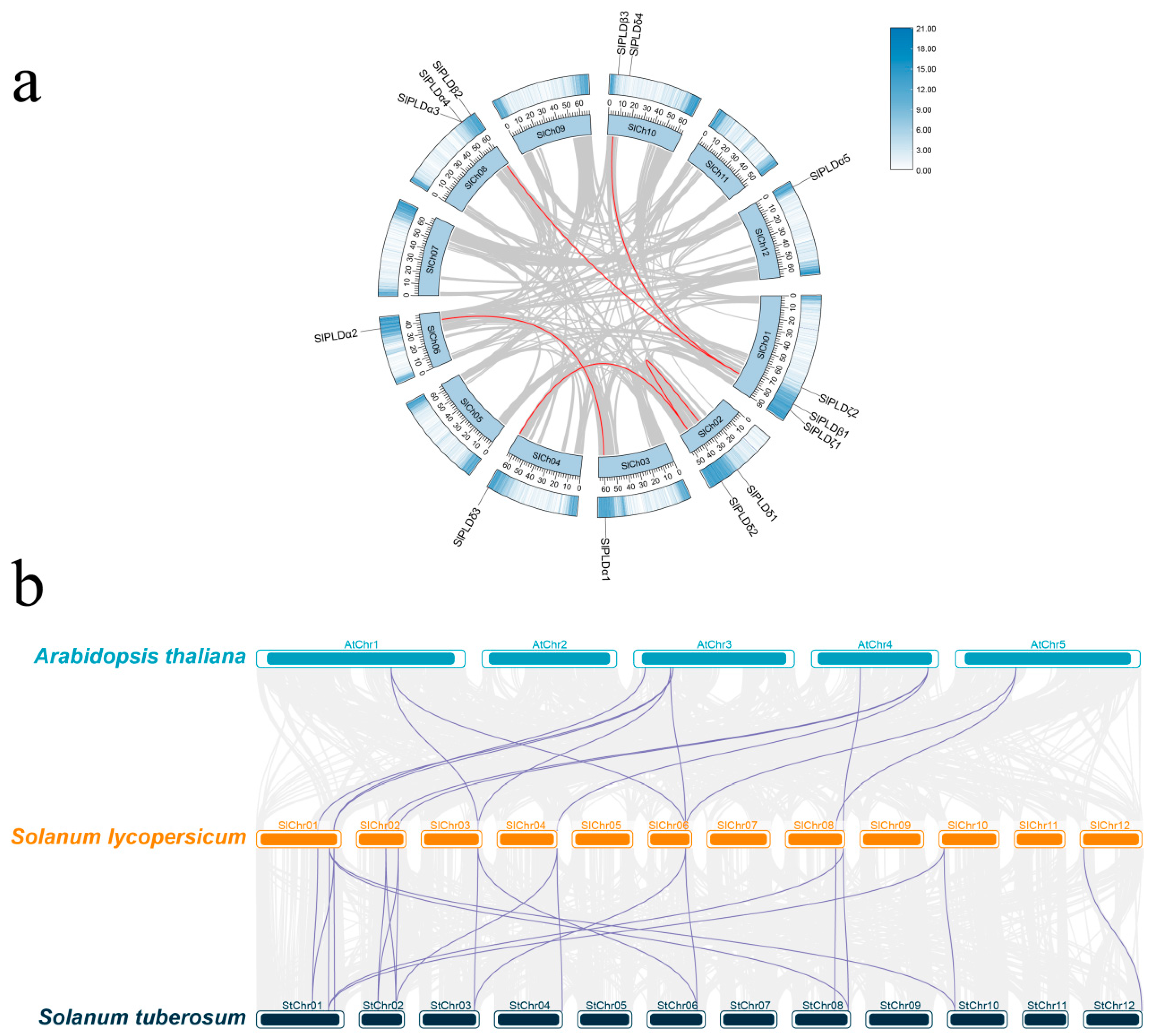
3.4. Chromosomal Distribution and Collinearity Analysis of SlPLD Genes
3.5. Cis-Elements in the Promoter of SlPLD Genes
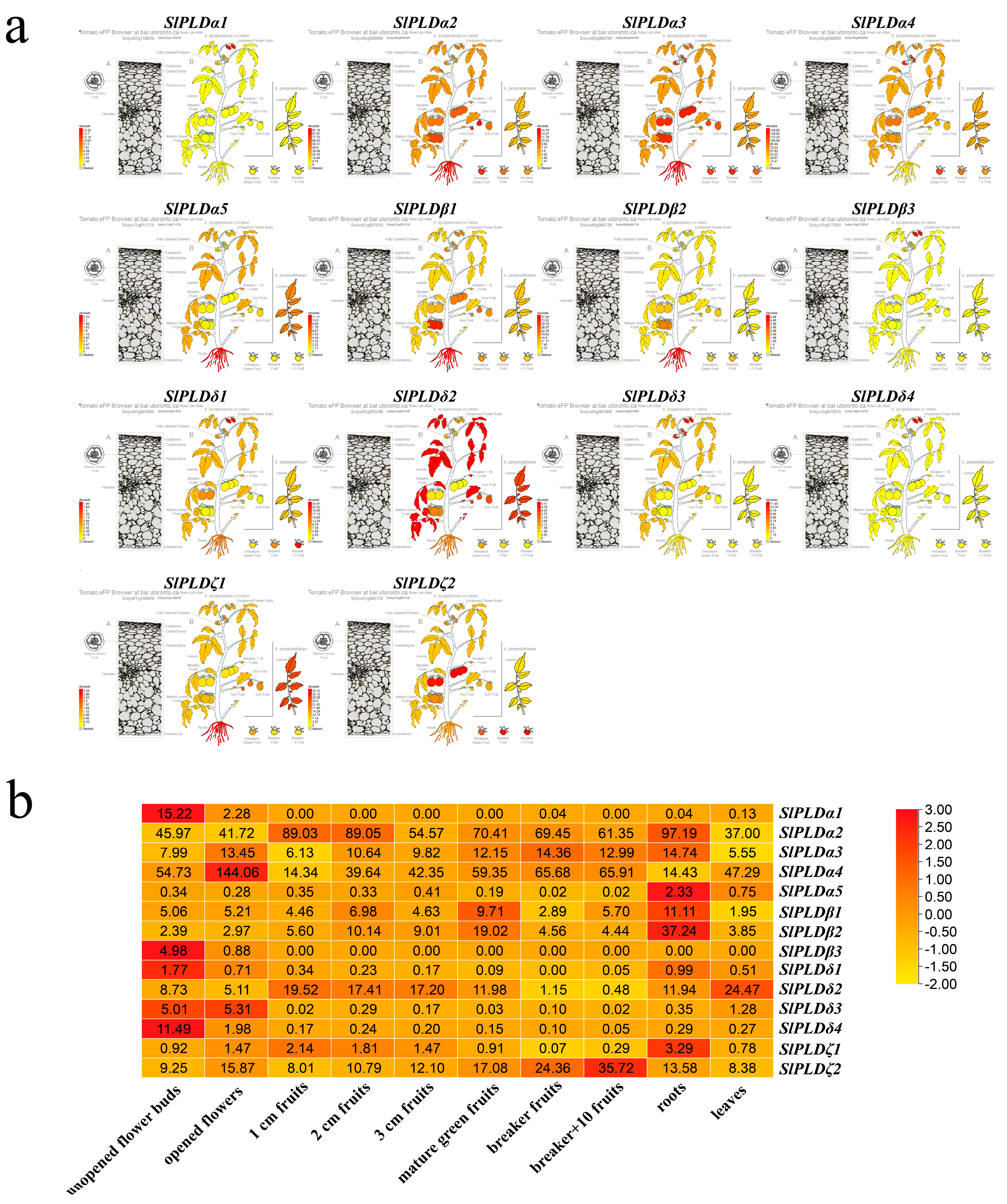
3.6. Expression Patterns of SlPLD Genes Revealed by Transcriptome Analysis
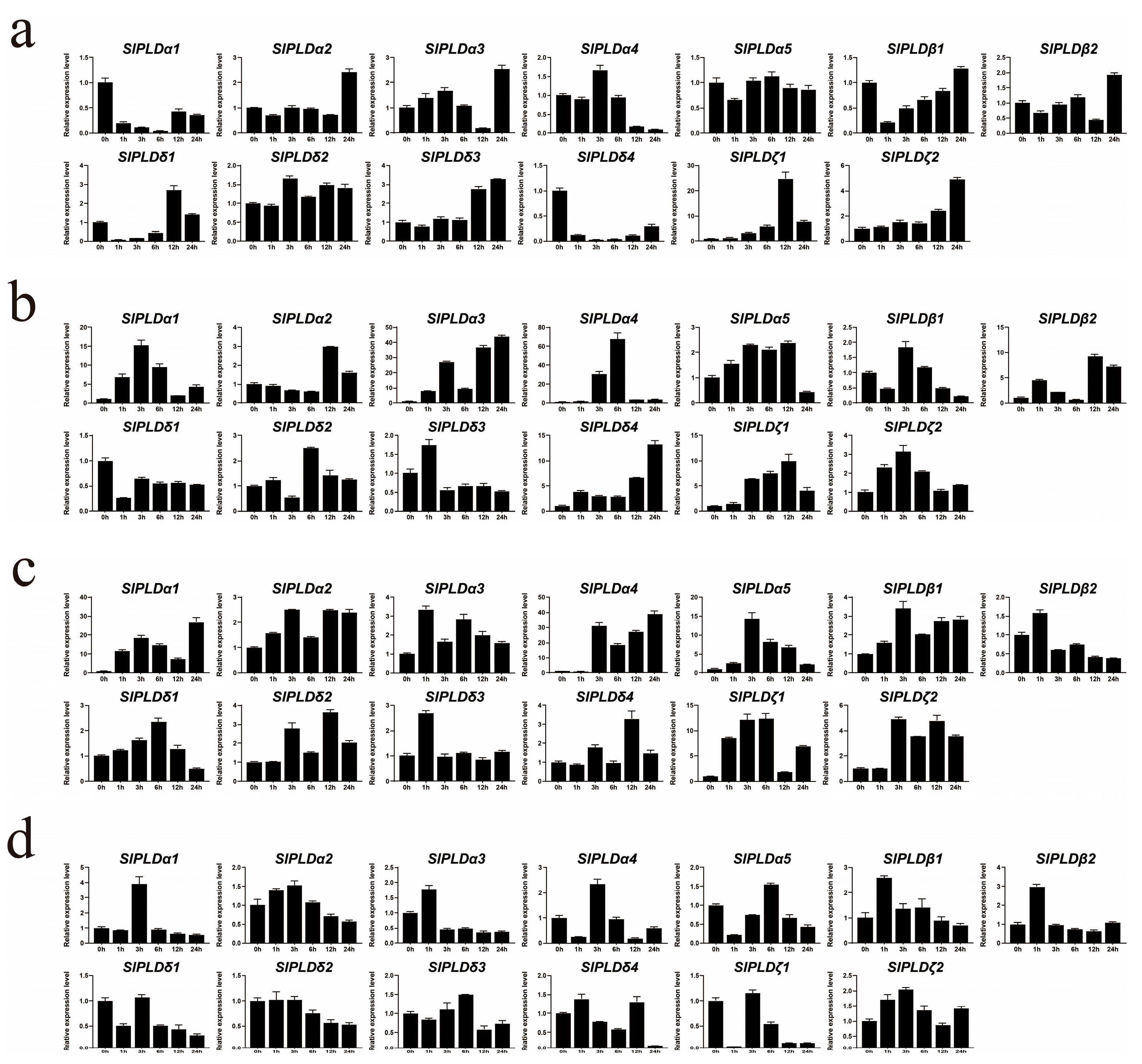
3.7. Expression Profiles of SlPLD Genes in Response to Abiotic Stress Exposure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.-C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant Phospholipases D and C and Their Diverse Functions in Stress Responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef]
- Wang, G.; Ryu, S.; Wang, X. Plant Phospholipases: An Overview. Methods Mol. Biol. 2012, 861, 123–137. [Google Scholar] [CrossRef]
- Wang, X. Plant Phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 211–231. [Google Scholar] [CrossRef]
- Jenkins, G.M.; Frohman, M.A. Phospholipase D: A Lipid Centric Review. Cell. Mol. Life Sci. 2005, 62, 2305–2316. [Google Scholar] [CrossRef] [PubMed]
- Onono, F.O.; Morris, A.J. Phospholipase D and Choline Metabolism. Lipid Signal. Hum. Dis. 2020, 259, 205–218. [Google Scholar] [CrossRef]
- Ufer, G.; Gertzmann, A.; Gasulla, F.; Röhrig, H.; Bartels, D. Identification and Characterization of the Phosphatidic Acid-Binding A. Thaliana Phosphoprotein PLDrp1 That Is Regulated by PLDα1 in a Stress-Dependent Manner. Plant J. 2017, 92, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Snyder, C.L.; Greer, M.S.; Weselake, R.J. Biology and Biochemistry of Plant Phospholipases. Crit. Rev. Plant Sci. 2011, 30, 239–258. [Google Scholar] [CrossRef]
- Wang, X.; Xu, L.; Zheng, L. Cloning and Expression of Phosphatidylcholine-Hydrolyzing Phospholipase D from Ricinus communis L. J. Biol. Chem. 1994, 269, 20312–20317. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Wang, X. The Arabidopsis Phospholipase D Family. Characterization of a Calcium-Independent and Phosphatidylcholine-Selective PLDζ1 with Distinct Regulatory Domains. Plant Physiol. 2002, 128, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Lin, F.; Xue, H.-W. Genome-Wide Analysis of the Phospholipase D Family in Oryza Sativa and Functional Characterization of PLDβ1 in Seed Germination. Cell Res. 2007, 17, 881–894. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhou, D.; Zhang, Q.; Zhang, W. Genomic Analysis of Phospholipase D Family and Characterization of GmPLDαs in Soybean (Glycine Max). J. Plant Res. 2012, 125, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cao, B.; Han, N.; Tao, Y.; Zhou, S.F.; Li, W.C.; Fu, F.L. Phospholipase D Family and Its Expression in Response to Abiotic Stress in Maize. Plant Growth Regul. 2017, 81, 197–207. [Google Scholar] [CrossRef]
- Lu, S.; Fadlalla, T.; Tang, S.; Li, L.; Ali, U.; Li, Q.; Guo, L. Genome-Wide Analysis of Phospholipase D Gene Family and Profiling of Phospholipids under Abiotic Stresses in Brassica Napus. Plant Cell Physiol. 2019, 60, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, C.; Zhang, M.; Yang, C.; Bao, Y.; Wang, D.; Chen, Q.; Chen, Y. Genome-Wide Analysis and Expression Profiling of the Phospholipase D Gene Family in Solanum Tuberosum. Biology 2021, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Shao, W.; Liu, X.; He, L.; Zhao, C.; Yu, G.; Xu, J. Genome-Wide Identification and Expression Analysis of Phospholipase D Gene in Leaves of Sorghum in Response to Abiotic Stresses. Physiol. Mol. Biol. Plants 2022, 28, 1261–1276. [Google Scholar] [CrossRef]
- Yuan, Y.; Yu, J.; Kong, L.; Zhang, W.; Hou, X.; Cui, G. Genome-Wide Investigation of the PLD Gene Family in Alfalfa (Medicago Sativa L.): Identification, Analysis and Expression. BMC Genom. 2022, 23, 243. [Google Scholar] [CrossRef]
- Bargmann, B.O.R.; Munnik, T. The Role of Phospholipase D in Plant Stress Responses. Curr. Opin. Plant Biol. 2006, 9, 515–522. [Google Scholar] [CrossRef]
- Cho, J.H.; Han, J.-S. Phospholipase D and Its Essential Role in Cancer. Mol. Cells 2017, 40, 805–813. [Google Scholar] [CrossRef]
- Eliáš, M.; Potocký, M.; Cvrčková, F.; Žárský, V. Molecular Diversity of Phospholipase D in Angiosperms. BMC Genom. 2002, 3, 2. [Google Scholar] [CrossRef]
- Li, J.; Yu, F.; Guo, H.; Xiong, R.; Zhang, W.; He, F.; Zhang, M.; Zhang, P. Crystal Structure of Plant PLDα1 Reveals Catalytic and Regulatory Mechanisms of Eukaryotic Phospholipase D. Cell Res. 2020, 30, 61–69. [Google Scholar] [CrossRef]
- Wang, X. Regulatory Functions of Phospholipase D and Phosphatidic Acid in Plant Growth, Development, and Stress Responses. Plant Physiol. 2005, 139, 566–573. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and Phosphatidic Acid Regulate NADPH Oxidase Activity and Production of Reactive Oxygen Species in ABA-Mediated Stomatal Closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef]
- Yu, H.Q.; Yong, T.M.; Li, H.J.; Liu, Y.P.; Zhou, S.F.; Fu, F.L.; Li, W.C. Overexpression of a Phospholipase Dα Gene from Ammopiptanthus Nanus Enhances Salt Tolerance of Phospholipase Dα1-Deficient Arabidopsis Mutant. Planta 2015, 242, 1495–1509. [Google Scholar] [CrossRef]
- Wang, J.; Ding, B.; Guo, Y.; Li, M.; Chen, S.; Huang, G.; Xie, X. Overexpression of a Wheat Phospholipase D Gene, TaPLDα, Enhances Tolerance to Drought and Osmotic Stress in Arabidopsis Thaliana. Planta 2014, 240, 103–115. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, X.; Wang, P.; Wang, H.; Ge, X.; Li, F.; Hou, Y. The Phospholipase D Gene GhPLDδ Confers Resistance to Verticillium Dahliae and Improves Tolerance to Salt Stress. Plant Sci. 2022, 321, 111322. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Zheng, S.; Li, W.; Huang, B.; Wang, X. Regulation of Plant Water Loss by Manipulating the Expression of Phospholipase Dα. Plant J. 2001, 28, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, Z.; Yue, Y.; Zhao, H.; Fei, X.; E., L.; Liu, C.; Chen, S.; Lai, J.; Song, W. Loss-of-Function Alleles of ZmPLD3 Cause Haploid Induction in Maize. Nat. Plants 2021, 7, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhu, H.; Lu, Y.; Zhu, H. The Regulation of Nutrient and Flavor Metabolism in Tomato Fruit. Veg. Res. 2022, 2, 5. [Google Scholar] [CrossRef]
- Kimura, S.; Sinha, N. Tomato (Solanum Lycopersicum): A Model Fruit-Bearing Crop. Cold Spring Harb. Protoc. 2008, 2008, pdb.emo105. [Google Scholar] [CrossRef] [PubMed]
- Bovy, A.; de Vos, R.; Kemper, M.; Schijlen, E.; Almenar Pertejo, M.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.; et al. High-Flavonol Tomatoes Resulting from the Heterologous Expression of the Maize Transcription Factor Genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [PubMed]
- Willits, M.G.; Kramer, C.M.; Prata, R.T.N.; De Luca, V.; Potter, B.G.; Steffens, J.C.; Graser, G. Utilization of the Genetic Resources of Wild Species To Create a Nontransgenic High Flavonoid Tomato. J. Agric. Food Chem. 2005, 53, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Laxalt, A.M.; Ter Riet, B.; Verdonk, J.C.; Parigi, L.; Tameling, W.I.L.; Vossen, J.; Haring, M.; Musgrave, A.; Munnik, T. Characterization of Five Tomato Phospholipase D cDNAs: Rapid and Specific Expression of LePLDβ1 on Elicitation with Xylanase. Plant J. 2001, 26, 237–247. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From Genotype to Phenotype to Breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Fei, Z.; Joung, J.-G.; Tang, X.; Zheng, Y.; Huang, M.; Lee, J.M.; McQuinn, R.; Tieman, D.M.; Alba, R.; Klee, H.J.; et al. Tomato Functional Genomics Database: A Comprehensive Resource and Analysis Package for Tomato Functional Genomics. Nucleic Acids Res. 2011, 39, D1156–D1163. [Google Scholar] [CrossRef]
- Mascia, T.; Santovito, E.; Gallitelli, D.; Cillo, F. Evaluation of Reference Genes for Quantitative Reverse-Transcription Polymerase Chain Reaction Normalization in Infected Tomato Plants. Mol. Plant Pathol. 2010, 11, 805–816. [Google Scholar] [CrossRef]
- Campos, M.L. A Novel Role for a Phospholipase D in Plant Immunity. Plant Physiol. 2020, 183, 33–34. [Google Scholar] [CrossRef]
- Li, J.; Wang, X. Phospholipase D and Phosphatidic Acid in Plant Immunity. Plant Sci. 2019, 279, 45–50. [Google Scholar] [CrossRef]
- Arhab, Y.; Abousalham, A.; Noiriel, A. Plant Phospholipase D Mining Unravels New Conserved Residues Important for Catalytic Activity. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2019, 1864, 688–703. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early Allopolyploid Evolution in the Post-Neolithic Brassica Napus Oilseed Genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Ohashi, Y.; Oka, A.; Rodrigues-Pousada, R.; Possenti, M.; Ruberti, I.; Morelli, G.; Aoyama, T. Modulation of Phospholipid Signaling by GLABRA2 in Root-Hair Pattern Formation. Science 2003, 300, 1427–1430. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Qin, C.; Welti, R.; Wang, X. Double Knockouts of Phospholipases Dζ1 and Dζ2 in Arabidopsis Affect Root Elongation during Phosphate-Limited Growth But Do Not Affect Root Hair Patterning. Plant Physiol. 2006, 140, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, B.O.R.; Laxalt, A.M.; ter Riet, B.; van Schooten, B.; Merquiol, E.; Testerink, C.; Haring, M.A.; Bartels, D.; Munnik, T. Multiple PLDs Required for High Salinity and Water Deficit Tolerance in Plants. Plant Cell Physiol. 2009, 50, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Deepika; Biswas, D.K.; Chandrasekar, R.; Singh, A. Genome-Wide Identification, Structure Analysis and Expression Profiling of Phospholipases D under Hormone and Abiotic Stress Treatment in Chickpea (Cicer Arietinum). Int. J. Biol. Macromol. 2021, 169, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, B.O.R.; Laxalt, A.M.; ter Riet, B.; Schouten, E.; Van Leeuwen, W.; Dekker, H.L.; De Koster, C.G.; Haring, M.A.; Munnik, T. LePLDβ1 Activation and Relocalization in Suspension-Cultured Tomato Cells Treated with Xylanase. Plant J. 2006, 45, 358–368. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Gene Locus | Protein Length | Molecular Weight | PI |
|---|---|---|---|---|---|
| SlPLDα1 | Solyc03g116620.3.1 | Chr03:60414826..60418303+ | 811 | 92,638.49 | 5.41 |
| SlPLDα2 | Solyc06g068090.3.1 | Chr06:39833679..39838185+ | 809 | 92,200.33 | 5.39 |
| SlPLDα3 | Solyc08g066790.4.1 | Chr08:53735254..53739172+ | 807 | 92,750.81 | 6.22 |
| SlPLDα4 | Solyc08g066800.4.1 | Chr08:53752165..53758183+ | 807 | 92,027.6 | 5.63 |
| SlPLDα5 | Solyc12g011170.3.1 | Chr12:4055864..4060853+ | 848 | 96,687.18 | 6.58 |
| SlPLDβ1 | Solyc01g091910.4.1 | Chr01:77680828..77694493+ | 1106 | 123,113.22 | 6.54 |
| SlPLDβ2 | Solyc08g080130.3.1 | Chr08:61577138..61583167+ | 846 | 95,641.76 | 6.75 |
| SlPLDβ3 | Solyc10g017650.3.1 | Chr10:5319066..5325156+ | 1106 | 124,037.59 | 7.24 |
| SlPLDδ1 | Solyc02g061850.4.1 | Chr02:31366227..31376103- | 850 | 97,673.09 | 6.67 |
| SlPLDδ2 | Solyc02g083340.4.1 | Chr02:44778224..44786418- | 866 | 98,649.5 | 6.87 |
| SlPLDδ3 | Solyc04g082000.4.1 | Chr04:63789988..63798274+ | 839 | 95,664.13 | 7.09 |
| SlPLDδ4 | Solyc10g024370.3.1 | Chr10:13291114..13296807+ | 839 | 94,118.42 | 8.44 |
| SlPLDζ1 | Solyc01g100020.4.1 | Chr01:82361097..82372211+ | 1052 | 120,394.27 | 6.08 |
| SlPLDζ2 | Solyc01g065720.4.1 | Chr01:65078959..65099137- | 1106 | 125,961.43 | 6.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Zhu, W.; Wang, F.; Wang, H. Genome-Wide Investigation of the PLD Gene Family in Tomato: Identification, Analysis, and Expression. Genes 2024, 15, 326. https://doi.org/10.3390/genes15030326
Guo X, Zhu W, Wang F, Wang H. Genome-Wide Investigation of the PLD Gene Family in Tomato: Identification, Analysis, and Expression. Genes. 2024; 15(3):326. https://doi.org/10.3390/genes15030326
Chicago/Turabian StyleGuo, Xudong, Wenying Zhu, Fu Wang, and Hui Wang. 2024. "Genome-Wide Investigation of the PLD Gene Family in Tomato: Identification, Analysis, and Expression" Genes 15, no. 3: 326. https://doi.org/10.3390/genes15030326
APA StyleGuo, X., Zhu, W., Wang, F., & Wang, H. (2024). Genome-Wide Investigation of the PLD Gene Family in Tomato: Identification, Analysis, and Expression. Genes, 15(3), 326. https://doi.org/10.3390/genes15030326





