Modeling within and between Sub-Genomes Epistasis of Synthetic Hexaploid Wheat for Genome-Enabled Prediction of Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic Evaluations
2.2. Genotyping
2.3. Phenotypic Model for Disease Traits TS, SNB and SB
2.4. Genomic Prediction Models
2.4.1. Whole Genome Models
2.4.2. Sub-Genome Models
2.5. Cross-Validation Schemes
2.6. Software
3. Results
3.1. Estimated Variance Components for the Different Traits and Statistical Models
3.2. Genomic Prediction
4. Discussion
4.1. Activation of Genes from Sub-Genome D with Those of Sub-Genome A and B
4.2. Study of Epistasis of Sub-Genome A, B, and D Genes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santantonio, N.; Jannink, J.L.; Sorrells, M. Prediction of subgenome additive and interaction effects in allohexaploid wheat. G3 Genes Genomes Genet. 2019, 9, 685–698. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Villareal, R.L.; Mujeeb-Kazi, A.; Fuentes-Davila, G.; Rajaram, S.; Del Toro, E. Resistance to Karnal Bunt (Tilletia indica Mitra) in Synthetic Hexaploid Wheats Derived from Triticum turgidum × T. tauschii. Plant Breed. 1994, 112, 63–69. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Hettel, G. (Eds.) Utilizing Wild Grass Biodiversity in Wheat Improvement: 15 Years of Wide Cross Research at CIMMYT; CIMMYT: Mexcio City, Mexcio, 1995. [Google Scholar]
- Mujeeb-Kazi, A.; Rosas, V.; Roldán, S. Conservation of the genetic variation of Triticum tauschii (Coss.) Schmalh. (Aegilops squarrosa auct. non L.) in synthetic hexaploid wheats (T. turgidum L. s. lat. 9 T. tauschii; 2n = 6x = 42, AABBDD) and its potential utilization for wheat improvement. Genet Resour. Crop. Evol. 1996, 43, 129–134. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Cano, S.; Rosas, V.; Cortes, A.; Delgado, R. Registration of five synthetic hexaploid wheat and seven bread wheat germplasm lines resistant to wheat spot blotch. Crop Sci. 2001, 41, 1653–1654. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Gul, A.; Ahmad, F.; Farooq, M.; Rizwan, S.; Bux, H.; Iftikhar, S.; Asad, S.; Delgado, R. Aegilops tauschii, as spot blotch (Cochliobolus sativus) resistance source for bread wheat improvement. Pak. J. Bot. 2007, 39, 1207–1216. [Google Scholar]
- Dreisigacker, S.; Kishii, M.; Lage, J.; Warburton, M. Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Aust. J. Agric. Res. 2008, 59, 413–420. [Google Scholar] [CrossRef]
- Rosyara, U.; Kishii, M.; Payne, T.; Sansaloni, C.P.; Singh, R.P.; Braun, H.-J.; Dreisigacker, S. Genetic Contribution of Synthetic Hexaploid Wheat to CIMMYT’s Spring Bread Wheat Breeding Germplasm. Sci. Rep. 2019, 9, 12355. [Google Scholar] [CrossRef]
- Aberkane, H.; Payne, T.; Kishi, M.; Smale, M.; Amri, A.; Jamora, N. Transferring diversity of goat grass to farmers’ fields through the development of synthetic hexaploid wheat. Food Secur. 2020, 12, 1017–1033. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Yang, J.; Zaitlen, N.A.; Goddard, M.E.; Visscher, P.M.; Price, A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014, 46, 100–106. [Google Scholar] [CrossRef]
- Gianola, D.; Fariello, M.I.; Naya, H.; Schön, C.C. Genome-wide association studies with a genomic relationship matrix: A case study with wheat and Arabidopsis. G3 Genes Genomes Genet. 2016, 6, 3241–3256. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; De Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Gholami, M.; Wimmer, V.; Sansaloni, C.; Petroli, C.; Hearne, S.J.; Covarrubias-Pazaran, G.; Rensing, S.; Heise, J.; Pérez-Rodríguez, P.; Dreisigacker, S.; et al. A comparison of the adoption of genomic selection across different breeding institutions. Front. Plant Sci. 2021, 12, 728567. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Bonnett, D.; Li, Y.; Crossa, J.; Dreisigacker, S.; Basnet, B.; Pérez-Rodríguez, P.; Alvarado, G.; Jannink, J.L.; Poland, J.; Sorrells, M. Response to Early Generation Genomic Selection for Yield in Wheat. Front. Plant Sci. 2022, 12, 718611. [Google Scholar] [CrossRef]
- Dreisigacker, S.; Martini, J.W.R.; Cuevas, J.; Pérez-Rodríguez, P.; Lozano-Ramírez, N.; Huerta, J.; Singh, P.K.; Crespo-Herrera, L.; Bentley, A.R.; Crossa, J. Genomic prediction of Synthetic Hexaploid Wheat upon tetraploid durum and diploid Aegilops parental pools. Plant Genome 2023, submitted.
- Crossa, J.; de los Campos, G.D.L.; Pérez, P.; Gianola, D.; Burgueño, J.; Araus, J.L.; Makumbi, D.; Singh, R.P.; Dreisigacker, S.; Yan, J.; et al. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 2010, 186, 713–724. [Google Scholar] [CrossRef]
- Jiang, Y.; Reif, J.C. Modeling epistasis in genomic selection. Genetics 2015, 201, 759–768. [Google Scholar] [CrossRef]
- Martini, J.W.R.; Wimmer, V.; Erbe, M.; Simianer, H. Epistasis and covariance: How gene interaction translates into genomic relationship. Theor. Appl. Genet. 2016, 129, 963–976. [Google Scholar] [CrossRef]
- Martini, J.W.R.; Toledo, F.H.; Crossa, J. On the approximation of interaction effect models by Hadamard powers of the additive genomic relationship. Theor. Popul. Biol. 2020, 132, 16–23. [Google Scholar] [CrossRef]
- Jiang, Y.; Reif, J.C. Efficient algorithms for calculating epistatic genomic relationship matrices. Genetics 2020, 216, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Ramírez, N.; Dreisigacker, S.; Sansaloni, C.P.; He, X.; Sandoval-Islas, J.S.; Pérez-Rodríguez, P.; Carballo, A.C.; Nava-Díaz, C.; Kishii, M.; Singh, P.K. Genome-Wide Association Study for Resistance to Tan Spot in Synthetic Hexaploid Wheat. Plants 2022, 11, 433. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Ramirez, N.; Dreisigacker, S.; Sansaloni, C.P.; He, X.; Sandoval-Islas, J.S.; Pérez-Rodríguez, P.; Carballo, A.C.; Diaz, C.N.; Kishii, M.; Singh, P.K. Genome-Wide Association Study for Spot Blotch Resistance in Synthetic Hexaploid Wheat. Genes 2022, 13, 1387. [Google Scholar] [CrossRef]
- Lamari, L.; Bernier, C.C. Evaluation of wheat lines and cultivars to tan spot [Pyrenophora tritici-repentis] based on lesion type. Can. J. Plant Pathol. 1989, 11, 49–56. [Google Scholar] [CrossRef]
- Dreisigacker, S.; Sehgal, D.; Jaimez, A.R.; Garrido, B.L.; Savala, S.M.; Núñez-Ríos, C.; Mollins, J.; Mall, S. CIMMYT Wheat Molecular Genetics: Laboratory Protocols and Applications to Wheat Breeding; Mollins, J., Mall, S., Eds.; CIMMYT: Mexico City, Mexico, 2016. [Google Scholar]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, P54. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Maccaferri, M.; Sanguineti, M.C.; Mantovani, P.; Demontis, A.; Massi, A.; Ammar, K.; Kolmer, J.A.; Czembor, J.H.; Ezrati, S.; Tuberosa, R. Association mapping of leaf rust response in durum wheat. Mol. Breed. 2019, 26, 189–228. [Google Scholar] [CrossRef]
- Luo, M.-C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Gianola, D. Theory and Analysiss of the Threshold Characters. J. Anim. Sci. 1982, 54, 1079–1096. [Google Scholar] [CrossRef]
- Lopez-Cruz, M.; Crossa, J.; Bonnett, D.; Dreisigacker, S.; Poland, J.; Jannink, J.L.; Singh, R.P.; Autrique, E.; de los Campos, G. Increased prediction accuracy in wheat breeding trials using a marker × environment interaction genomic selection model. G3 2015, 5, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Basnet, B.R.; Crossa, J.; Dreisigacker, S.; Perez-Rodriguez, P.; Manes, Y.; Singh, R.P.; Royara, U.R.; Camarillo-Castillo, F.; Murua, M. Hybrid Wheat prediction using genomic, pedigree and environmental covariables interaction models. Plant Genome 2019, 12, 180051. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; de los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.G.; Friesen, T.L.; Xu, S.S.; Faris, J.D. Identification of novel tan spot resistance loci beyond the known host- selective toxin insensitivity genes in wheat. Theor. Appl. Genet. 2008, 117, 873–881. [Google Scholar] [CrossRef]
- Akdemir, D.; Jannink, J.L. Locally epistatic genomic relationship matrices for genomic association and prediction. Genetics 2015, 199, 857–871. [Google Scholar] [CrossRef]
- Vitezica, Z.; Legarra, A.; Toro, M.A.; Varona, L. Orthogonal estimates of variances for additive, dominance, and epistatic effects in population. Genetics 2017, 206, 1297–1307. [Google Scholar] [CrossRef]
- González-Diéguez, D.; Crossa, J.; Rivera-Amado, C.; Pinera-Chavez, F.; Pinto, F.; Pierre, C.S.; Pérez-Rodríguez, P.; Dreisigacker, S.; Reynolds, M. Genome-based predictions of sub-genome genetic interactions effects in wheat populations. In Proceedings of the XVIIIth Eucarpia Biometrics in Plant Breeding Conference, Paris, France, 21–23 September 2022. [Google Scholar]
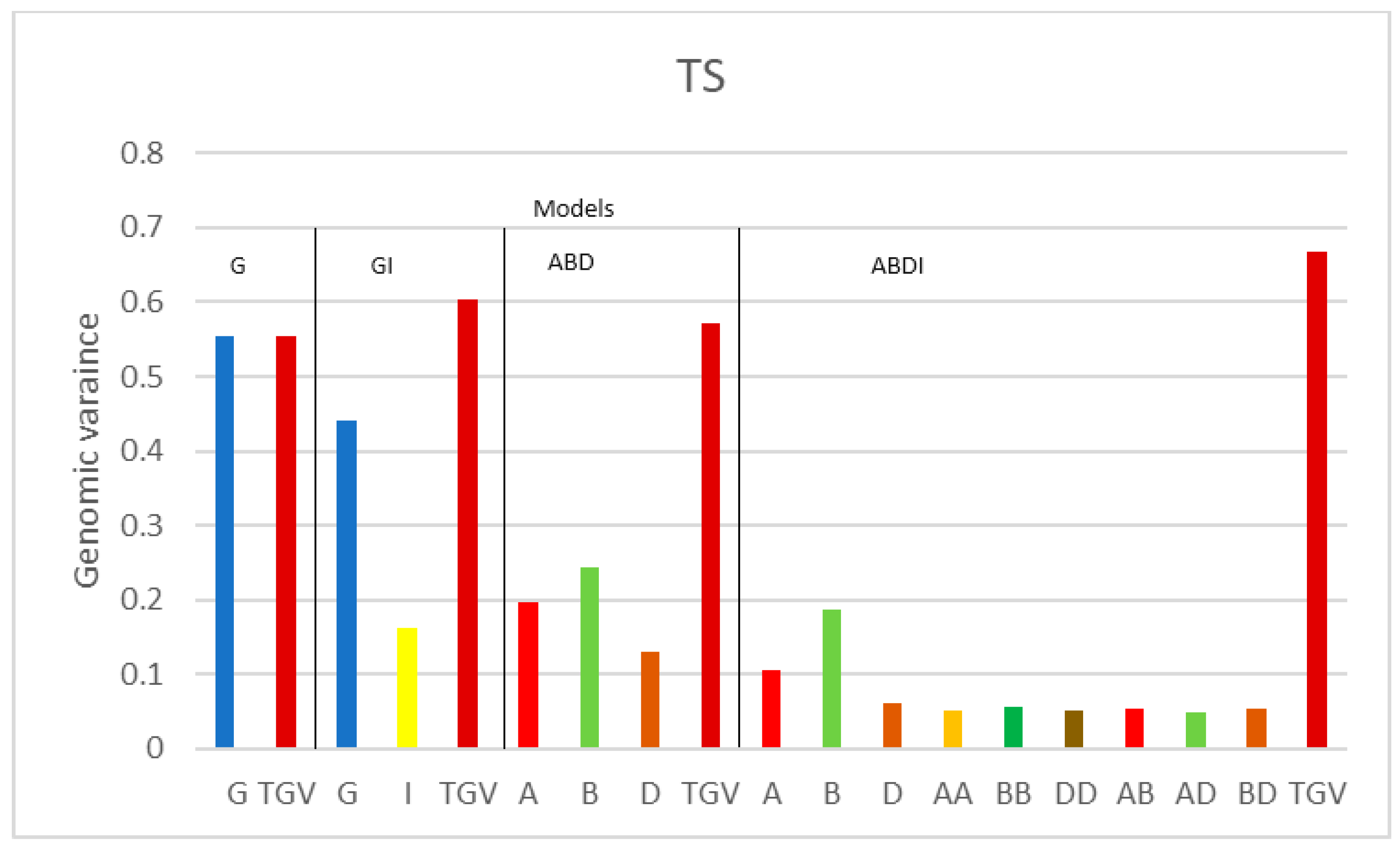

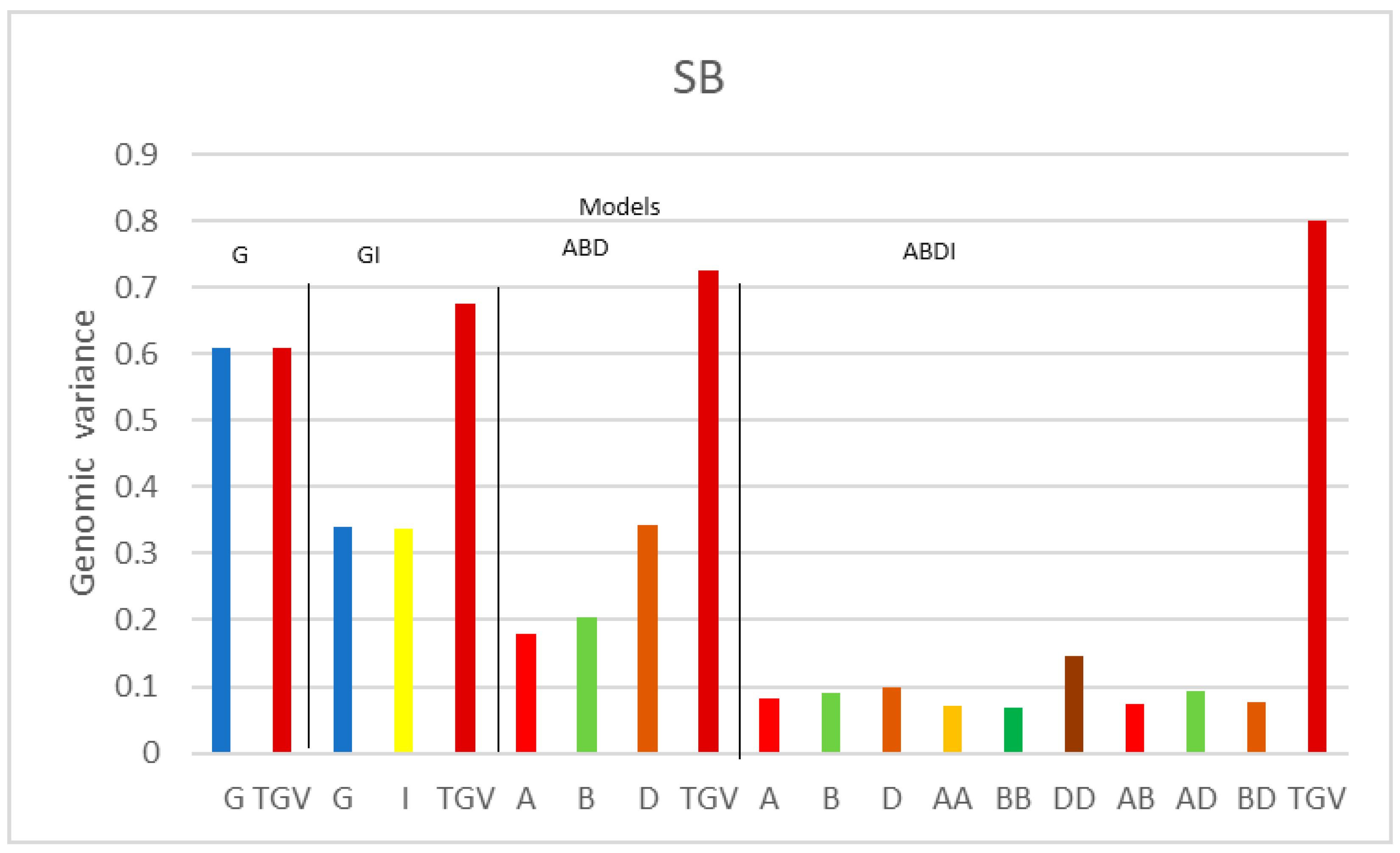

| Variance Components | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRAIT | Model | Res | |||||||||||
| TS | G | 0.555 | 0.330 | ||||||||||
| GI | 0.440 | 0.163 | 0.261 | ||||||||||
| ABD | 0.196 | 0.244 | 0.131 | 0.326 | |||||||||
| ABDI | 0.105 | 0.187 | 0.061 | 0.052 | 0.056 | 0.052 | 0.053 | 0.048 | 0.054 | 0.234 | |||
| SNB | G | 0.724 | 0.403 | ||||||||||
| GI | 0.386 | 0.383 | 0.259 | ||||||||||
| ABD | 0.321 | 0.176 | 0.265 | 0.396 | |||||||||
| ABDI | 0.148 | 0.091 | 0.072 | 0.067 | 0.068 | 0.109 | 0.065 | 0.100 | 0.091 | 0.235 | |||
| SB | G | 0.608 | 0.535 | ||||||||||
| GI | 0.339 | 0.337 | 0.416 | ||||||||||
| ABD | 0.179 | 0.205 | 0.343 | 0.503 | |||||||||
| ABDI | 0.081 | 0.090 | 0.100 | 0.072 | 0.069 | 0.146 | 0.075 | 0.092 | 0.077 | 0.353 | |||
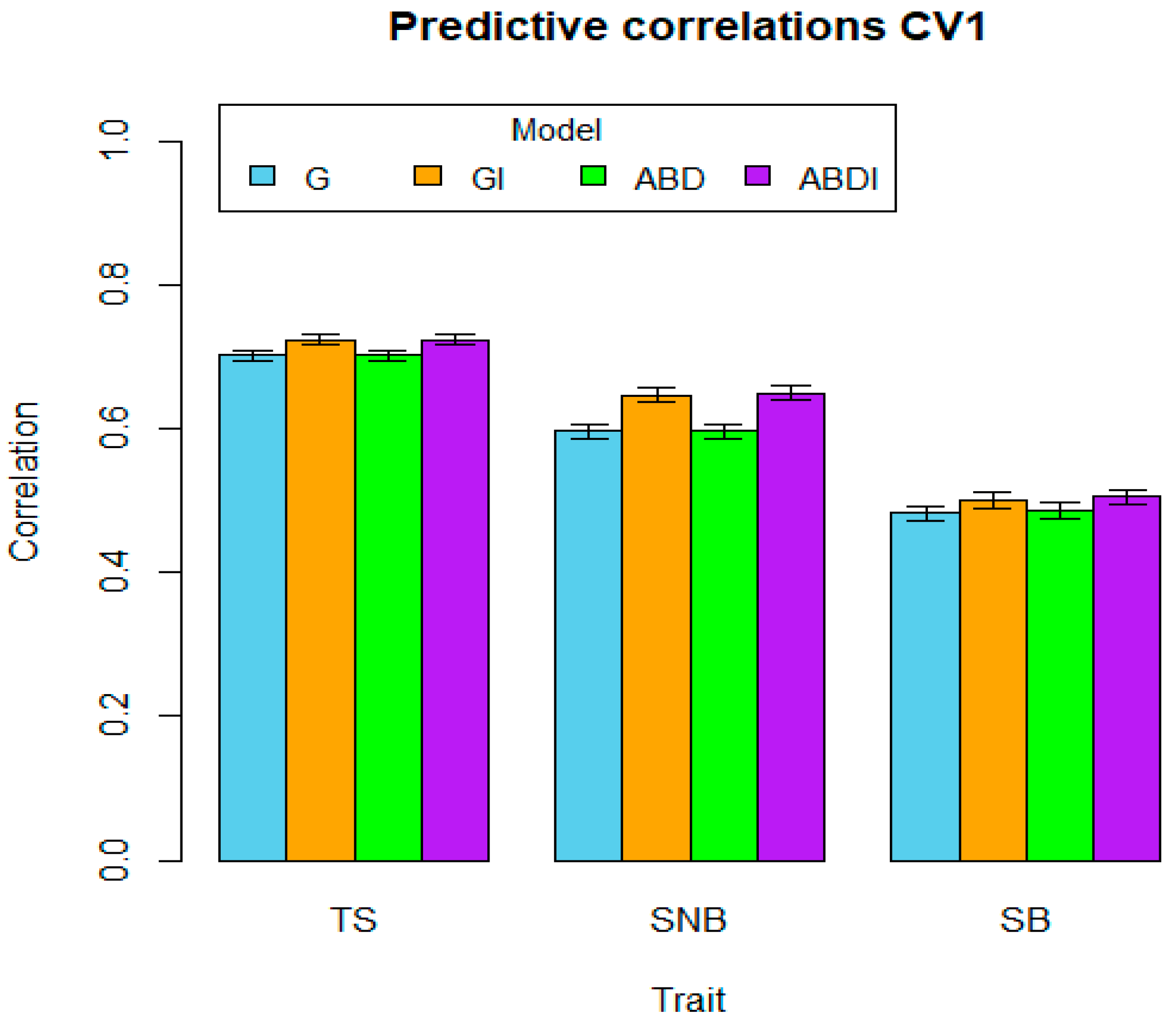
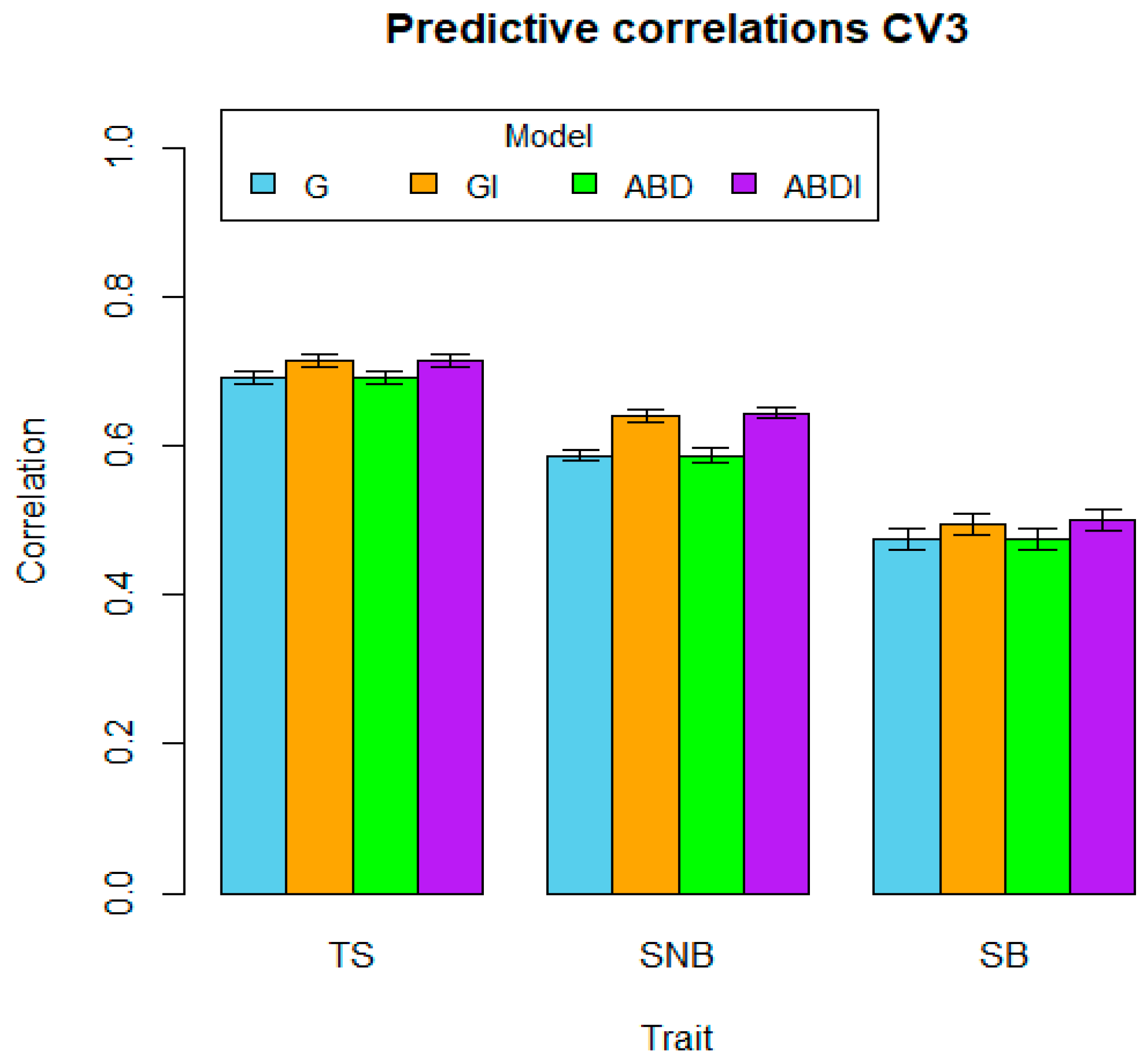
| Traits | CV1 | CV2 | CV3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | GI | ABD | ABDI | G | GI | ABD | ABDI | G | GI | ABD | ABDI | ||
| TS | Mean | 0.702 | 0.724 | 0.702 | 0.724 | 0.632 | 0.639 | 0.632 | 0.635 | 0.691 | 0.714 | 0.691 | 0.715 |
| SE | 0.007 | 0.007 | 0.007 | 0.007 | 0.010 | 0.011 | 0.010 | 0.011 | 0.009 | 0.009 | 0.009 | 0.009 | |
| SNB | Mean | 0.597 | 0.647 | 0.596 | 0.650 | 0.481 | 0.487 | 0.479 | 0.484 | 0.587 | 0.641 | 0.587 | 0.644 |
| SE | 0.010 | 0.009 | 0.009 | 0.009 | 0.017 | 0.017 | 0.017 | 0.017 | 0.008 | 0.008 | 0.009 | 0.008 | |
| SB | Mean | 0.482 | 0.500 | 0.486 | 0.506 | 0.370 | 0.394 | 0.371 | 0.401 | 0.475 | 0.495 | 0.476 | 0.500 |
| SE | 0.011 | 0.011 | 0.011 | 0.010 | 0.017 | 0.015 | 0.017 | 0.014 | 0.014 | 0.014 | 0.014 | 0.014 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuevas, J.; González-Diéguez, D.; Dreisigacker, S.; Martini, J.W.R.; Crespo-Herrera, L.; Lozano-Ramirez, N.; Singh, P.K.; He, X.; Huerta, J.; Crossa, J. Modeling within and between Sub-Genomes Epistasis of Synthetic Hexaploid Wheat for Genome-Enabled Prediction of Diseases. Genes 2024, 15, 262. https://doi.org/10.3390/genes15030262
Cuevas J, González-Diéguez D, Dreisigacker S, Martini JWR, Crespo-Herrera L, Lozano-Ramirez N, Singh PK, He X, Huerta J, Crossa J. Modeling within and between Sub-Genomes Epistasis of Synthetic Hexaploid Wheat for Genome-Enabled Prediction of Diseases. Genes. 2024; 15(3):262. https://doi.org/10.3390/genes15030262
Chicago/Turabian StyleCuevas, Jaime, David González-Diéguez, Susanne Dreisigacker, Johannes W. R. Martini, Leo Crespo-Herrera, Nerida Lozano-Ramirez, Pawan K. Singh, Xinyao He, Julio Huerta, and Jose Crossa. 2024. "Modeling within and between Sub-Genomes Epistasis of Synthetic Hexaploid Wheat for Genome-Enabled Prediction of Diseases" Genes 15, no. 3: 262. https://doi.org/10.3390/genes15030262
APA StyleCuevas, J., González-Diéguez, D., Dreisigacker, S., Martini, J. W. R., Crespo-Herrera, L., Lozano-Ramirez, N., Singh, P. K., He, X., Huerta, J., & Crossa, J. (2024). Modeling within and between Sub-Genomes Epistasis of Synthetic Hexaploid Wheat for Genome-Enabled Prediction of Diseases. Genes, 15(3), 262. https://doi.org/10.3390/genes15030262







