Molecular Phylogenetics and Mitochondrial Genomic Evolution in the Endemic Genus Pielomastax (Orthoptera: Eumastacoidea) in China
Abstract
1. Introduction
2. Materials and Methods
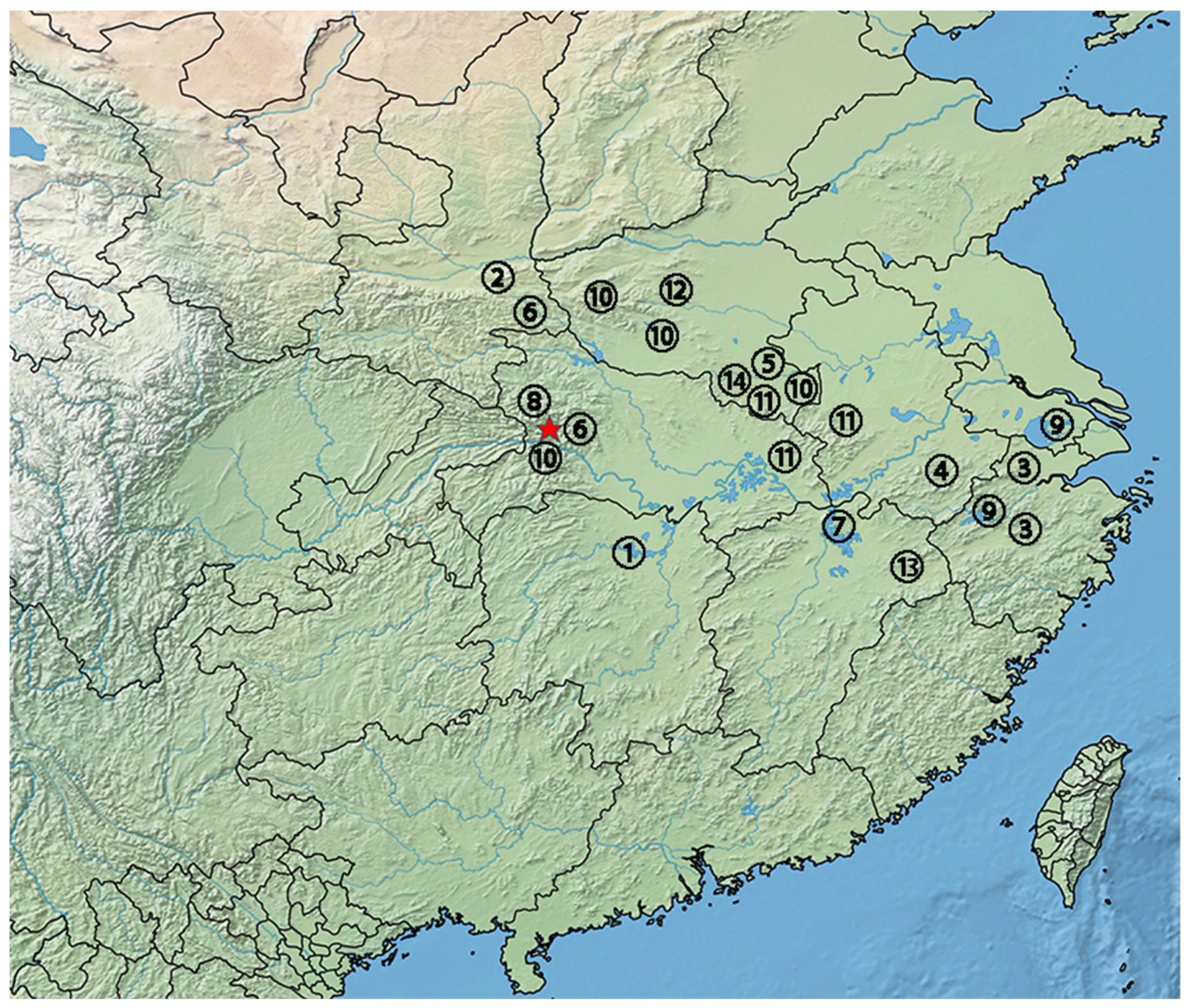
2.1. Specimen Collection, Morphological Data Acquisition and Mitogenome Sequencing
| Family | Subfamily | Species | Accession Number | Elevation (m) | Location | Reference |
|---|---|---|---|---|---|---|
| Chorotypidae | Episactinae | Pielomastax shennongjiaensis | PQ325292 | 784.3 | Shenlongjia, Hubei, China | This study |
| Pielomastax tenuicerca | PQ325293 | 1201.4 | Sanmenxia, Henan, China | This study | ||
| Pielomastax sp. | PQ325294 | 1042.2 | Shenlongjia, Hubei, China | This study | ||
| Pielomastax zhengi | JF411955 | [19] | ||||
| Pielomastax soochowensis | KM102728 | [20] | ||||
| Erianthinae | Chorotypus fenestratus | KM657339 | [21] | |||
| Chininae | China manfispoides | OQ241410 | [22] | |||
| Eumastacidae | Gomphomastacinae | Phytomastax pentaspinula | OQ241413 | [22] | ||
| Paramastacinae | Paramastax nigra | JX913772 | [23] | |||
| Episactidae | Episactinae | Erianthus versicolor | OQ241411 | [22] | ||
| Thericleidae | Thericleinae | Pseudothericles compressifrons | NC028061 | [21] | ||
| Tetrigidae | Tetrix japonica * | NC018543 | [24] | |||
| Tanaoceridae | Tanaocerus koebelei * | JX913774 | [23] | |||
| Pyrgomorphidae | Atractomorpha sinensis * | EU263919 | [25] | |||
| Acrididae | Locusta migratoria * | NC001712 | [26] |
2.2. Phylogenetic Reconstruction
2.3. Estimating Divergence Time and Comparative Phylogenetic Analyses
3. Results
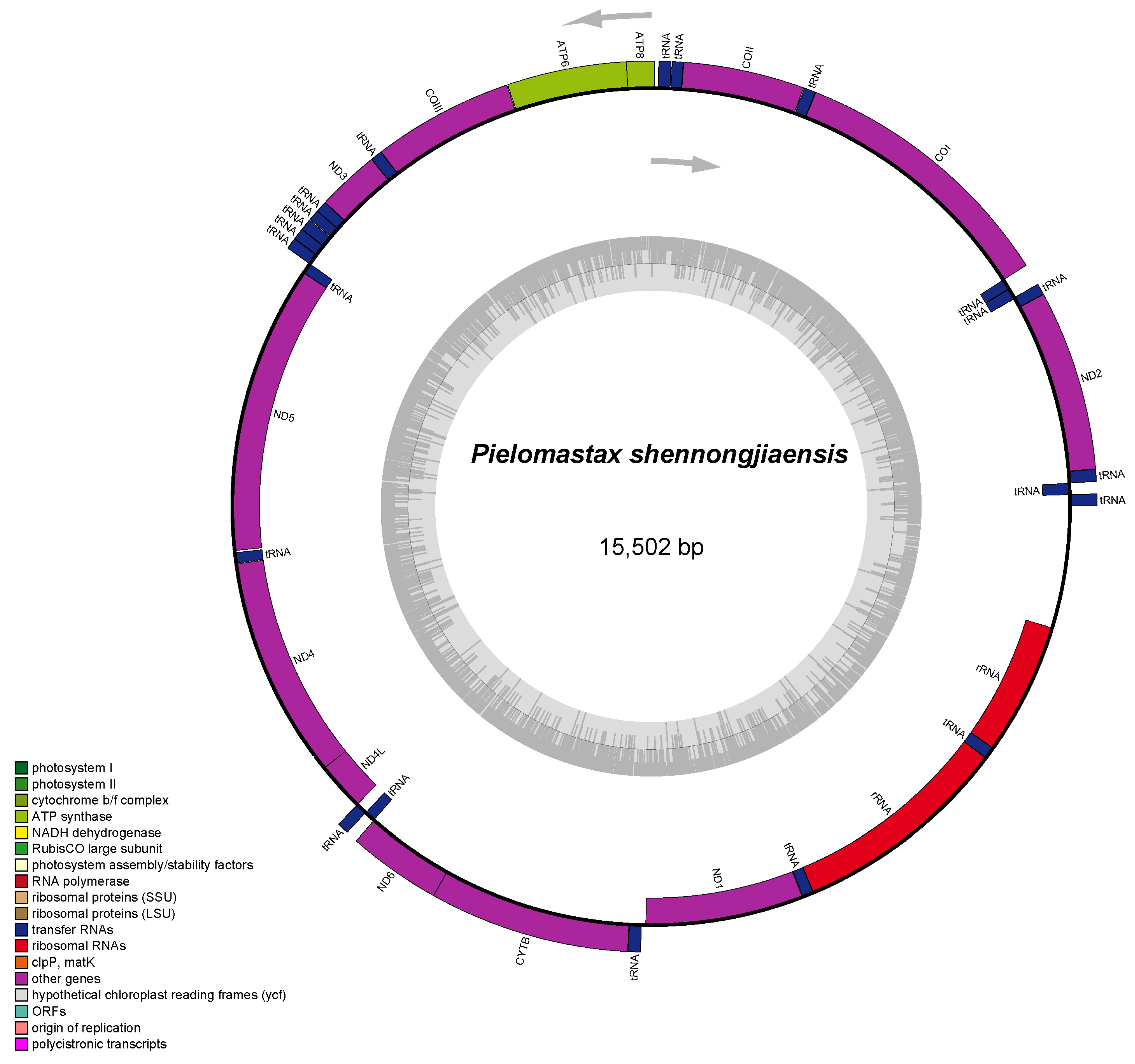
3.1. Mitochondrial Genome Assembly and Annotation
3.2. Phylogenetic Reconstruction
3.3. Divergence Time Estimation and PGLS Analysis
4. Discussion
4.1. Insect Mitochondrial Genome
4.2. Phylogenetic Position of Pielomastax in Eumastacoidea
4.3. Diversification within Episactinae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cigliano, M.M.; Braun, H.; Eades, D.C.; Otte, D. Orthoptera Species File. 2024. Available online: https://orthoptera.speciesfile.org/ (accessed on 13 July 2024).
- Amedegnato, C. African-American relationships in the acridians (Insecta, Orthoptera). In The Africa-South America Connection; Oxford Academic: Oxford, UK, 1993; pp. 59–75. [Google Scholar]
- Chang, K. Notes on the Eumastacinae (Orthoptera, Acrididae) from China with description of one new genus and two new species. Notes Ent Chin. Mus Heude 1937, 4, 35–46. [Google Scholar]
- Lin, L.; Zheng, Z.; Yang, R.; Xu, S. A review of the genus Pielomastax Chang (Orthoptera: Eumastacoidea) from China with description of a new species. Neotrop. Entomol. 2014, 43, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Fenn, J.D.; Song, H.; Cameron, S.L.; Whiting, M.F. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol. Phylogenet. Evol. 2008, 49, 59–68. [Google Scholar] [CrossRef]
- De Mandal, S.; Chhakchhuak, L.; Gurusubramanian, G.; Kumar, N.S. Mitochondrial markers for identification and phylogenetic studies in insects–A Review. DNA Barcodes 2014, 2, 1–9. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, Y.; Li, C.; Li, L.; Men, X. Mitochondrial DNA as a molecular marker in insect ecology: Current status and future prospects. Ann. Entomol. Soc. Am. 2021, 114, 470–476. [Google Scholar] [CrossRef]
- Da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, 1–22. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Q.; Lu, C.; Deng, J.; Huang, X. The first complete mitochondrial genome of Lachninae species and comparative genomics provide new insights into the evolution of gene rearrangement and the repeat region. Insects 2021, 12, 55. [Google Scholar] [CrossRef]
- Ye, F.; Li, H.; Xie, Q. Mitochondrial genomes from two specialized subfamilies of reduviidae (Insecta: Hemiptera) reveal novel gene rearrangements of true bugs. Genes 2021, 12, 1134. [Google Scholar] [CrossRef]
- Saenz Manchola, O.F.; Virrueta Herrera, S.; D’Alessio, L.M.; Yoshizawa, K.; García Aldrete, A.N.; Johnson, K.P. Mitochondrial genomes within bark lice (Insecta: Psocodea: Psocomorpha) reveal novel gene rearrangements containing phylogenetic signal. Syst. Entomol. 2021, 46, 938–951. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Yang, H.; Huang, Y. Analysis of the complete mitochondrial genome sequence of Pielomastax zhengi. Zool. Res. 2011, 32, 353–362. [Google Scholar]
- Bläser, M.; Predel, R. Evolution of neuropeptide precursors in Polyneoptera (Insecta). Front. Endocrinol. 2020, 11, 197. [Google Scholar] [CrossRef]
- Song, H.; Amédégnato, C.; Cigliano, M.M.; Desutter-Grandcolas, L.; Heads, S.W.; Huang, Y.; Otte, D.; Whiting, M.F. 300 million years of diversification: Elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics 2015, 31, 621–651. [Google Scholar] [CrossRef]
- Ge, J.J.; Ying, H.F.; Xu, S.Q.; Huang, H.T. Mitochondrial genome phylogeny reveals the deep-time origin of Gomphomastacinae (Orthoptera: Eumastacidae) and its alpine genera in China. J. Syst. Evol. 2023, 61, 1047–1055. [Google Scholar] [CrossRef]
- Leavitt, J.R.; Hiatt, K.D.; Whiting, M.F.; Song, H. Searching for the optimal data partitioning strategy in mitochondrial phylogenomics: A phylogeny of Acridoidea (Insecta: Orthoptera: Caelifera) as a case study. Mol. Phylogenet. Evol. 2013, 67, 494–508. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Feng, X.; Miao, W.-J.; Jiang, G.-F. The complete mitochondrial genome of grouse locust Tetrix japonica (Insecta: Orthoptera: Tetrigoidea). Mitochondrial DNA 2012, 23, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.-M.; Shi, H.-W.; Huang, Y. Complete mitochondrial genome and secondary structures of lrRNA and srRNA of Atractomorpha sinensis (Orthoptera, Pyrgomorphidae). Zool. Res. 2007, 28, 580–588. [Google Scholar]
- Flook, P.; Rowell, C.; Gellissen, G. The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. J. Mol. Evol. 1995, 41, 928–941. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.-I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Dowling, D.K.; Wolff, J.N. Evolutionary genetics of the mitochondrial genome: Insights from Drosophila. Genetics 2023, 224, iyad036. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Norman, H.; Creedy, T.J.; Jordaens, K.; Moran, K.M.; Young, A.; Mengual, X.; Skevington, J.H.; Vogler, A.P. The phylogeny and evolutionary ecology of hoverflies (Diptera: Syrphidae) inferred from mitochondrial genomes. Mol. Phylogenet. Evol. 2023, 184, 107759. [Google Scholar] [CrossRef] [PubMed]
- Antil, S.; Abraham, J.S.; Sripoorna, S.; Maurya, S.; Dagar, J.; Makhija, S.; Bhagat, P.; Gupta, R.; Sood, U.; Lal, R. DNA barcoding, an effective tool for species identification: A review. Mol. Biol. Rep. 2023, 50, 761–775. [Google Scholar] [CrossRef]
- Akter, K.; Meem, S.; Islam, S.; Mamun, M.; Abedin, M.; Borsha, F.; Ghosh, A.; Hossain, M.M. Utilizing COI gene for the Identification of thirteen Hesperiidae butterflies and determining their genetic relationship. J. Biodivers. Conserv. Bioresour. Manag. 2023, 9, 21–28. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, S.; Xu, J.; Guo, Y.; Yang, G. Adaptive evolution of mitochondrial energy metabolism genes associated with increased energy demand in flying insects. PLoS ONE 2014, 9, e99120. [Google Scholar] [CrossRef]
- Kang, N.; Hu, H. Adaptive evidence of mitochondrial genes in Pteromalidae and Eulophidae (Hymenoptera: Chalcidoidea). PLoS ONE 2023, 18, e0294687. [Google Scholar] [CrossRef]
- Yang, M.R.; Zhou, Z.J.; Chang, Y.L.; Zhao, L.H. The mitochondrial genome of the quiet-calling katydids, Xizicus fascipes (Orthoptera: Tettigoniidae: Meconematinae). J. Genet. 2012, 91, 141–153. [Google Scholar] [CrossRef]
- Łukasik, P.; Chong, R.A.; Nazario, K.; Matsuura, Y.; Bublitz, D.A.C.; Campbell, M.A.; Meyer, M.C.; Van Leuven, J.T.; Pessacq, P.; Veloso, C. One hundred mitochondrial genomes of cicadas. J. Hered. 2019, 110, 247–256. [Google Scholar] [CrossRef]
- Mao, S.; Yuan, H.; Chang, H.; Shi, F.; Zhou, Y. Comparative mitochondrial genomics of Shoveliteratura triangula (Orthoptera, Tettigoniidae, Meconematinae) and the first description of a female specimen. Zootaxa 2020, 4751, 507–520. [Google Scholar] [CrossRef]
- Cantatore, P.; Gadaleta, M.; Roberti, M.; Saccone, C.; Wilson, A. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature 1987, 329, 853–855. [Google Scholar] [CrossRef]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, L.; Liao, C.-Q.; Wang, X.; Wang, M.; Huang, G.-H. Comparative mitochondrial genome analysis and phylogenetic relationship among lepidopteran species. Gene 2022, 830, 146516. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, Y. Characterization of two complete mitochondrial genomes of Ledrinae (Hemiptera: Cicadellidae) and phylogenetic analysis. Insects 2020, 11, 609. [Google Scholar] [CrossRef] [PubMed]
- Matt, S.; Flook, P.; Rowell, C. A partial molecular phylogeny of the Eumastacoidea s. lat. (Orthoptera, Caelifera). J. Orthoptera Res. 2008, 17, 43–55. [Google Scholar] [CrossRef]
- Zhenhan, W.; Barosh, P.J.; Zhonghai, W.; Daogong, H.; Xun, Z.; Peisheng, Y. Vast early Miocene lakes of the central Tibetan Plateau. Geol. Soc. Am. Bull. 2008, 120, 1326–1337. [Google Scholar] [CrossRef]
- Jin, H.; Chang, X.; Wang, S. Evolution of permafrost on the Qinghai-Xizang (Tibet) Plateau since the end of the late Pleistocene. J. Geophys. Res. Earth Surf. 2007, 112, F2. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.-H.; Zhang, K.; Xu, S.-Q.; Ding, Y. Molecular Phylogenetics and Mitochondrial Genomic Evolution in the Endemic Genus Pielomastax (Orthoptera: Eumastacoidea) in China. Genes 2024, 15, 1260. https://doi.org/10.3390/genes15101260
Lu J-H, Zhang K, Xu S-Q, Ding Y. Molecular Phylogenetics and Mitochondrial Genomic Evolution in the Endemic Genus Pielomastax (Orthoptera: Eumastacoidea) in China. Genes. 2024; 15(10):1260. https://doi.org/10.3390/genes15101260
Chicago/Turabian StyleLu, Jun-Hui, Keyao Zhang, Sheng-Quan Xu, and Ying Ding. 2024. "Molecular Phylogenetics and Mitochondrial Genomic Evolution in the Endemic Genus Pielomastax (Orthoptera: Eumastacoidea) in China" Genes 15, no. 10: 1260. https://doi.org/10.3390/genes15101260
APA StyleLu, J.-H., Zhang, K., Xu, S.-Q., & Ding, Y. (2024). Molecular Phylogenetics and Mitochondrial Genomic Evolution in the Endemic Genus Pielomastax (Orthoptera: Eumastacoidea) in China. Genes, 15(10), 1260. https://doi.org/10.3390/genes15101260






