Integrating External Controls by Regression Calibration for Genome-Wide Association Study
Abstract
1. Introduction
2. Materials and Methods
- Step 1. Adjusting the Genotypes of External Controls by Regression Calibration
- (1)
- Without loss of generality, is assumed. A total of individuals with genotypes is chosen from external control samples.
- (2)
- A linear regression model is assumed for , where is the least-square estimate of .
- (3)
- (1) and (2) are repeated times. are obtained and the average value is calculated. Let for . When , let be 0, where is determined such that the frequency of 0 in the internal control genotypes is equal to the frequency of 0 in for . When , let be 1, where is determined such that the frequency of 1 in the internal control genotypes is equal to the frequency of 1 in for . When , let be 2.
- Step 2. Single-Variant Association Test
- Step 3. Calibrating Single-Variant Test Using the SPA and ER Methods
- (1). SPA Method
- (2). ER Method
3. Simulations
4. Result
4.1. Type I Error Rates
4.2. Power
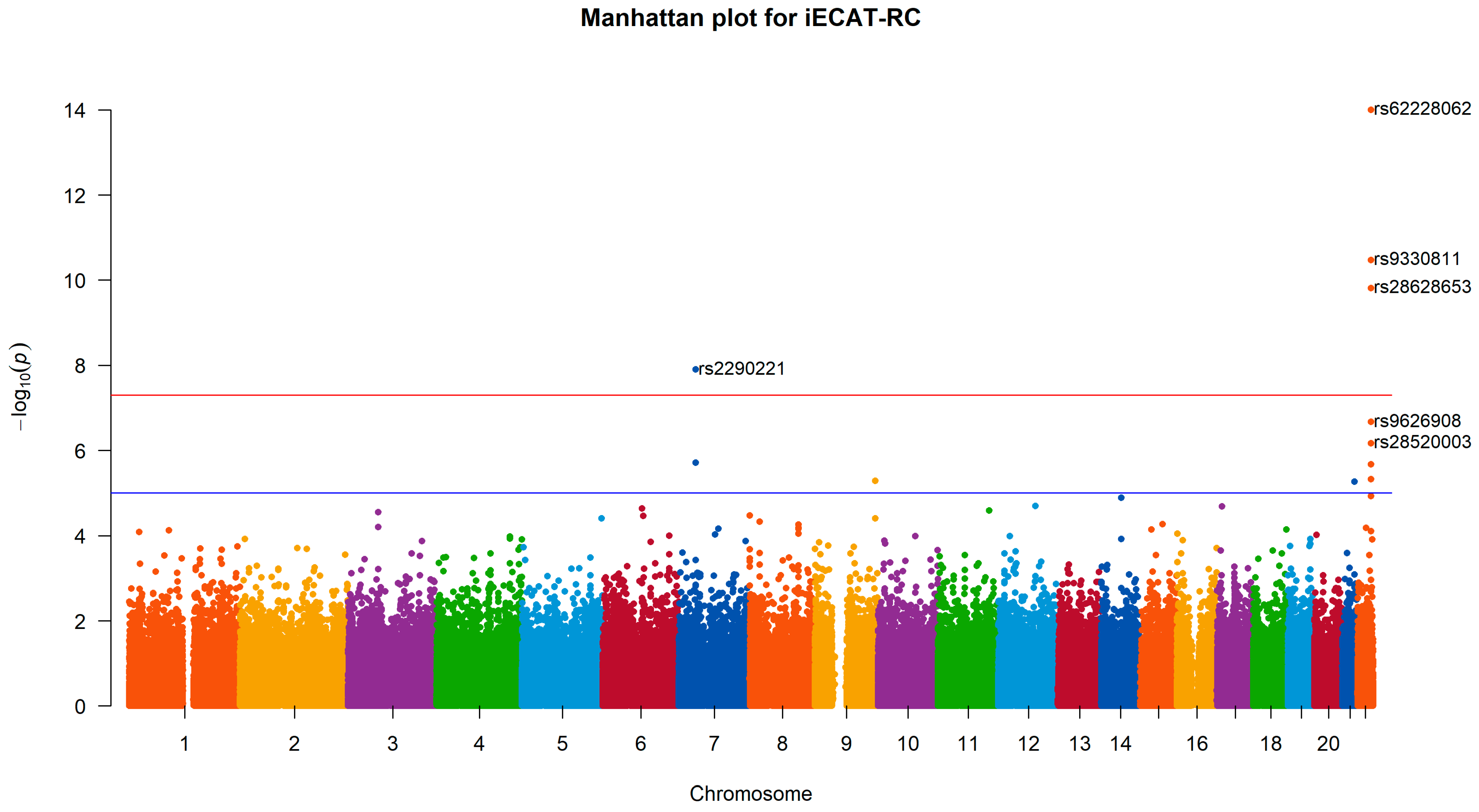
4.3. Application to the UK Biobank Data
5. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Price, A.L.; Spencer, C.C.; Donnelly, P. Progress and promise in understanding the genetic basis of common diseases. Proc. R. Soc. B Biol. Sci. 2015, 282, 20151684. [Google Scholar] [CrossRef]
- Sha, Q.; Wang, X.; Wang, X.; Zhang, S. Detecting association of rare and common variants by testing an optimally weighted combination of variants. Genet. Epidemiol. 2012, 36, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, S.; Sha, Q. Literature reviews on methods for rare variant association studies. Hum. Genet. Embryol. 2016, 6, 1000133. [Google Scholar]
- Homann, J.; Osburg, T.; Ohlei, O.; Dobricic, V.; Deecke, L.; Bos, I.; Vandenberghe, R.; Gabel, S.; Scheltens, P.; Teunissen, C.E.; et al. Genome-wide association study of Alzheimer’s disease brain imaging biomarkers and neuropsychological phenotypes in the European medical information framework for Alzheimer’s disease multimodal biomarker discovery dataset. Front. Aging Neurosci. 2022, 14, 840651. [Google Scholar] [CrossRef]
- Lin, D.Y.; Tang, Z.Z. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 2011, 89, 354–367. [Google Scholar] [CrossRef]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Skotte, L.; Korneliussen, T.S.; Albrechtsen, A. Association testing for next-generation sequencing data using score statistics. Genet. Epidemiol. 2012, 36, 430–437. [Google Scholar] [CrossRef]
- Marchini, J.; Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010, 11, 499–511. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.; Fuchsberger, C. Improving power for rare-variant tests by integrating external controls. Genet. Epidemiol. 2017, 41, 610–619. [Google Scholar] [CrossRef]
- Widmayer, S.J.; Evans, K.S.; Zdraljevic, S.; Andersen, E.C. Evaluating the power and limitations of genome-wide association studies in Caenorhabditis elegans. G3 2022, 12, jkac114. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.J.; Leal, S.M. SEQCHIP: A powerful method to integrate sequence and genotype data for the detection of rare variant associations. Bioinformatics 2012, 28, 1745–1751. [Google Scholar] [CrossRef][Green Version]
- Derkach, A.; Chiang, T.; Gong, J.; Addis, L.; Dobbins, S.; Tomlinson, I.; Houlston, R.; Pal, D.K.; Strug, L.J. Association analysis using next-generation sequence data from publicly available control groups: The robust variance score statistic. Bioinformatics 2014, 30, 2179–2188. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X. Analysis in case–control sequencing association studies with different sequencing depths. Biostatistics 2020, 21, 577–593. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, A.E.; Billups, S.C.; Pike, H.N.; Farooqi, I.S.; Zeggini, E.; Santorico, S.A.; Barroso, I.; Dupuis, J. ProxECAT: Proxy External Controls Association Test. A new case-control gene region association test using allele frequencies from public controls. PLoS Genet. 2018, 14, e1007591. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lee, S. Integrating external controls in case–control studies improves power for rare-variant tests. Genet. Epidemiol. 2022, 46, 145–158. [Google Scholar] [CrossRef]
- Dey, R.; Schmidt, E.M.; Abecasis, G.R.; Lee, S. A fast and accurate algorithm to test for binary phenotypes and its application to PheWAS. Am. J. Hum. Genet. 2017, 101, 37–49. [Google Scholar] [CrossRef]
- Lee, S.; Fuchsberger, C.; Kim, S.; Scott, L. An efficient resampling method for calibrating single and gene-based rare variant association analysis in case–control studies. Biostatistics 2016, 17, 1–5. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Li, Y.; Lee, S. Novel score test to increase power in association test by integrating external controls. Genet. Epidemiol. 2021, 45, 293–304. [Google Scholar] [CrossRef]
- Lee, S.; Abecasis, G.R.; Boehnke, M.; Lin, X. Rare-variant association analysis: Study designs and statistical tests. Am. J. Hum. Genet. 2014, 95, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Blackwell, T.; Boehnke, M.; Scott, L.J.; GoT2D Investigators. Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet. Epidemiol. 2013, 37, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- McGuirl, M.R.; Smith, S.P.; Sandstede, B.; Ramachandran, S. Detecting shared genetic architecture among multiple phenotypes by hierarchical clustering of gene-level association statistics. Genetics 2020, 215, 511–529. [Google Scholar] [CrossRef]
- Zhao, Z.; Bi, W.; Zhou, W.; VandeHaar, P.; Fritsche, L.G.; Lee, S. UK Biobank whole-exome sequence binary phenome analysis with robust region-based rare-variant test. Am. J. Hum. Genet. 2020, 106, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Paul, J.S.; Albrechtsen, A.; Song, Y.S. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011, 12, 443–451. [Google Scholar] [CrossRef]
- Tängdén, T.; Gustafsson, S.; Rao, A.S.; Ingelsson, E. A genome-wide association study in a large community-based cohort identifies multiple loci associated with susceptibility to bacterial and viral infections. Sci. Rep. 2022, 12, 2582. [Google Scholar] [CrossRef]
- Hu, Y.J.; Liao, P.; Johnston, H.R.; Allen, A.S.; Satten, G.A. Testing rare-variant association without calling genotypes allows for systematic differences in sequencing between cases and controls. PLoS Genet. 2016, 12, e1006040. [Google Scholar] [CrossRef]
- Liang, X.; Cao, X.; Sha, Q.; Zhang, S. HCLC-FC: A novel statistical method for phenome-wide association studies. PLoS ONE 2022, 17, e0276646. [Google Scholar] [CrossRef]
- Green, H.D.; Jones, A.; Evans, J.P.; Wood, A.R.; Beaumont, R.N.; Tyrrell, J.; Frayling, T.M.; Smith, C.; Weedon, M.N. A genome-wide association study identifies 5 loci associated with frozen shoulder and implicates diabetes as a causal risk factor. PLoS Genet. 2021, 17, e1009577. [Google Scholar] [CrossRef] [PubMed]
- Michou, L.; Lermusiaux, J.L.; Teyssedou, J.P.; Bardin, T.; Beaudreuil, J.; Petit-Teixeira, E. Genetics of Dupuytren’s disease. Jt. Bone Spine 2012, 79, 7–12. [Google Scholar] [CrossRef]
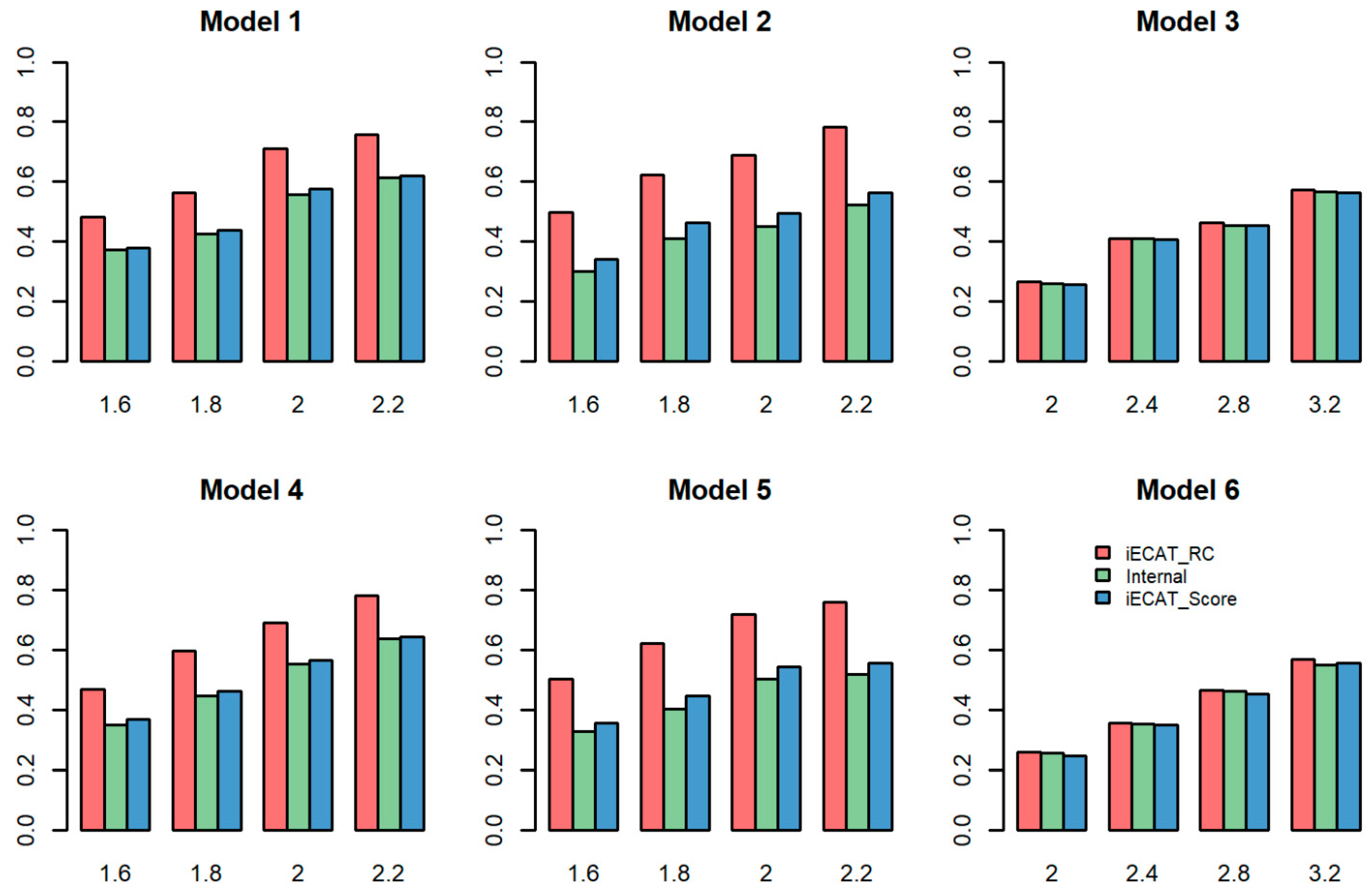
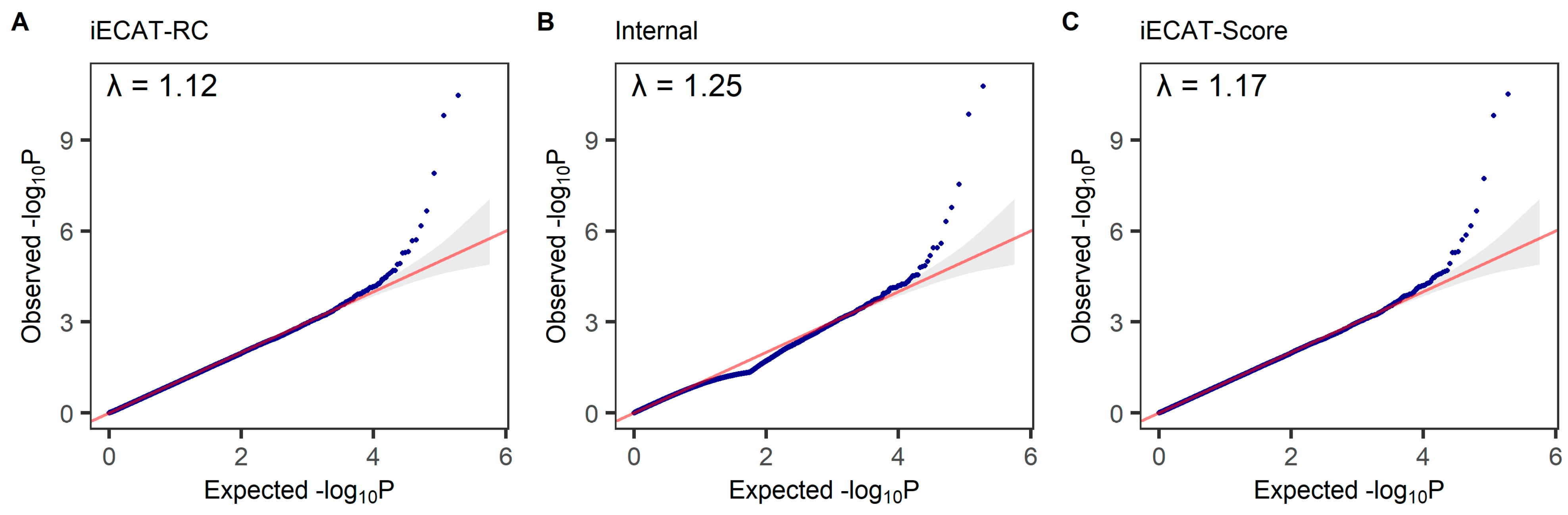
| Model | Significance Level | iECAT-RC | iECAT-N | Internal | iECAT-Score |
|---|---|---|---|---|---|
| Model 1 | 0.05 | 0.0382 | 0.3956 | 0.0512 | 0.0482 |
| 0.01 | 0.0057 | 0.3352 | 0.0102 | 0.0096 | |
| 0.001 | 3.00 × 10−4 | 0.2771 | 0.001 | 0.001 | |
| 1 × 10−4 | 1.00 × 10−4 | 0.2429 | 1.00 × 10−4 | 0 | |
| Model 2 | 0.05 | 0.0397 | 0.4163 | 0.0348 | 0.0394 |
| 0.01 | 0.0078 | 0.3685 | 0.0087 | 0.0089 | |
| 0.001 | 9.00 × 10−4 | 0.3263 | 4.00 × 10−4 | 0.0013 | |
| 1 × 10−4 | 1.00 × 10−4 | 0.2919 | 0 | 2.00 × 10−4 | |
| Model 3 | 0.05 | 0.0457 | 0.113 | 0.0136 | 0.0357 |
| 0.01 | 0.0111 | 0.0628 | 0.004 | 0.0081 | |
| 0.001 | 6.00 × 10−4 | 0.0345 | 5.00 × 10−4 | 3.00 × 10−4 | |
| 1 × 10−4 | 0 | 0.0223 | 0 | 0 | |
| Model 4 | 0.05 | 0.0372 | 0.4269 | 0.0511 | 0.0475 |
| 0.01 | 0.0065 | 0.3513 | 0.0105 | 0.0101 | |
| 0.001 | 4.00 × 10−4 | 0.2804 | 9.00 × 10−4 | 0.001 | |
| 1 × 10−4 | 0 | 0.2359 | 3.00 × 10−4 | 1.00 × 10−4 | |
| Model 5 | 0.05 | 0.0494 | 0.457 | 0.0335 | 0.0446 |
| 0.01 | 0.0107 | 0.3876 | 0.0079 | 0.0096 | |
| 0.001 | 0.0017 | 0.3244 | 9.00 × 10−4 | 0.001 | |
| 1 × 10−4 | 4.00 × 10−4 | 0.2806 | 0 | 1.00 × 10−4 | |
| Model 6 | 0.05 | 0.0467 | 0.1013 | 0.0133 | 0.0342 |
| 0.01 | 0.011 | 0.0569 | 0.0042 | 0.007 | |
| 0.001 | 0.0012 | 0.0291 | 9.00 × 10−4 | 7.00 × 10−4 | |
| 1 × 10−4 | 1.00 × 10−4 | 0.0169 | 0 | 0 |
| Study | Samples Size | ||
|---|---|---|---|
| Cases | Controls | Totals | |
| UKBL (internal) | 229 | 22,472 | 22,701 |
| UKBB (external) | 297,068 | 297,068 | |
| Total | 229 | 318,540 | 319,769 |
| Chromosome | SNP | Base Position | Genes | iECAT-RC | iECAT-Score | Internal |
|---|---|---|---|---|---|---|
| 7 | rs2290221 | 37987632 | SFRP4, EPDR1 | 1.26 × 10−8 | 2.91 × 10−8 | 1.86 × 10−8 |
| 22 | rs9330811 | 46362396 | WNT7B | 1.65 × 10−11 | 3.37 × 10−11 | 3.00 × 10−11 |
| 22 | rs62228062 | 46381234 | WNT7B | 6.04 × 10−18 | 8.82 × 10−18 | 6.04 × 10−18 |
| 22 | rs28628653 | 46396925 | LOC730668 | 1.54 × 10−10 | 1.40 × 10−10 | 1.54 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Yan, S.; Cao, X.; Zhang, S.; Sha, Q. Integrating External Controls by Regression Calibration for Genome-Wide Association Study. Genes 2024, 15, 67. https://doi.org/10.3390/genes15010067
Zhu L, Yan S, Cao X, Zhang S, Sha Q. Integrating External Controls by Regression Calibration for Genome-Wide Association Study. Genes. 2024; 15(1):67. https://doi.org/10.3390/genes15010067
Chicago/Turabian StyleZhu, Lirong, Shijia Yan, Xuewei Cao, Shuanglin Zhang, and Qiuying Sha. 2024. "Integrating External Controls by Regression Calibration for Genome-Wide Association Study" Genes 15, no. 1: 67. https://doi.org/10.3390/genes15010067
APA StyleZhu, L., Yan, S., Cao, X., Zhang, S., & Sha, Q. (2024). Integrating External Controls by Regression Calibration for Genome-Wide Association Study. Genes, 15(1), 67. https://doi.org/10.3390/genes15010067






