Spontaneous Emergence of Multicellular Heritability
Abstract
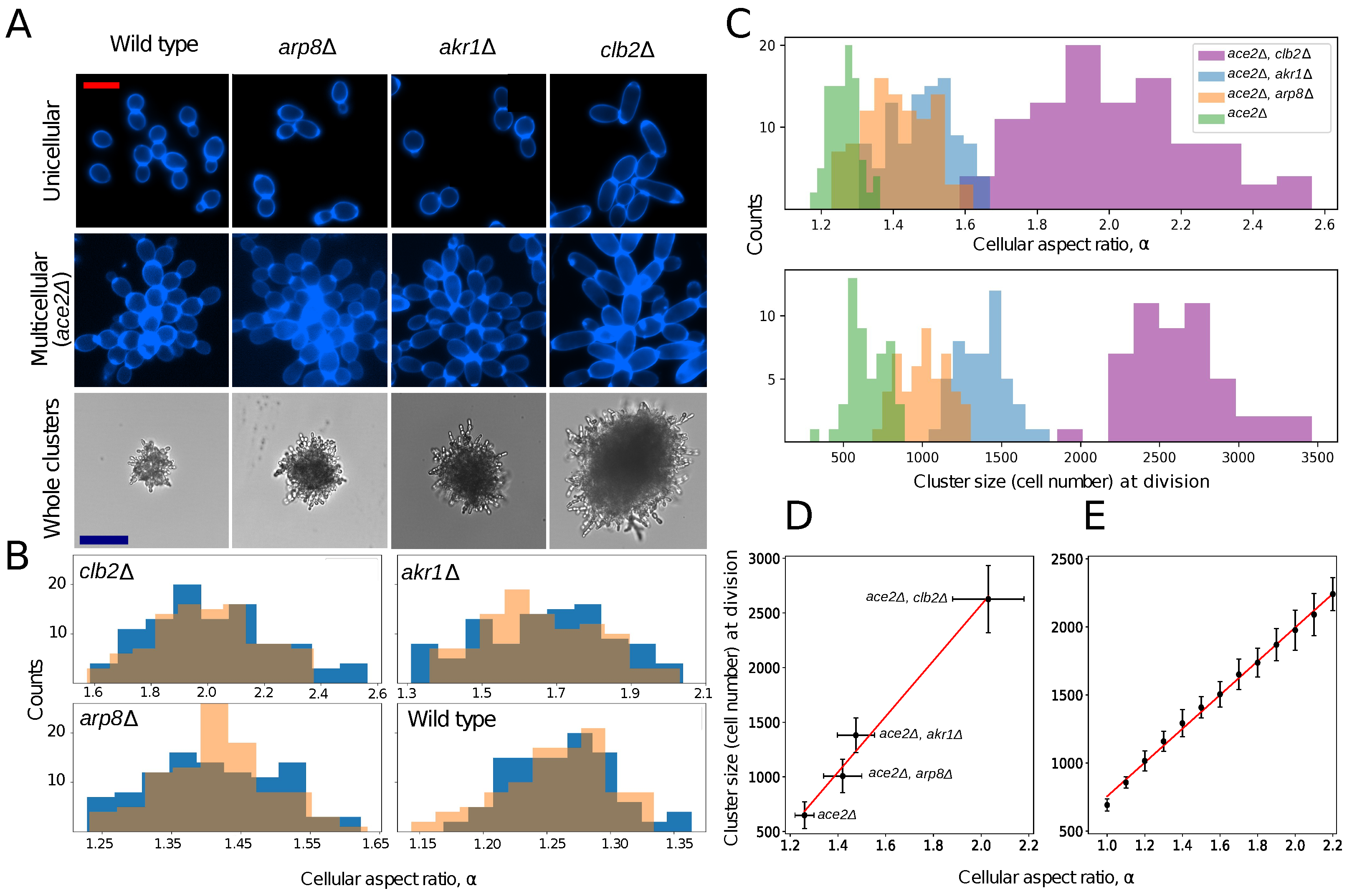
:1. Introduction
2. Results
2.1. Experimental System
2.2. Quantifying Heritability
3. Discussion
4. Methods
4.1. Strain Construction
4.2. Cell Culture
4.3. Measuring the Cellular Packing Fraction
4.4. Quantifying Cellular Aspect Ratio
4.5. Quantifying Cluster Size at Division
4.6. Calculating Heritability by Partitioning Variance
4.7. Biophysical Simulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abedin, M.; King, N. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 2010, 20, 734–742. [Google Scholar] [CrossRef]
- Tarnita, C.E.; Taubes, C.H.; Nowak, M.A. Evolutionary construction by staying together and coming together. J. Theor. Biol. 2013, 320, 10–22. [Google Scholar] [CrossRef]
- Niklas, K.J.; Newman, S.A. The origins of multicellular organisms. Evol. Dev. 2013, 15, 41–52. [Google Scholar] [CrossRef]
- Kapsetaki, S.E.; Fisher, R.M.; West, S.A. Predation and the formation of multicellular groups in algae. Evol. Ecol. Res. 2016, 17, 651–669. [Google Scholar]
- Iwasa, K.; Murakami, S. Palmelloid Formation of Chlamydomonas: I. Palmelloid Induction by Organic Acids. Physiol. Plant. 1968, 21, 1224–1233. [Google Scholar] [CrossRef]
- Alegado, R.A.; Brown, L.W.; Cao, S.; Dermenjian, R.K.; Zuzow, R.; Fairclough, S.R.; Clardy, J.; King, N. A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. eLife 2012, 1, e00013. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F.; Borrello, M.; Travisano, M. Experimental evolution of multicellularity. Proc. Natl. Acad. Sci. USA 2012, 109, 1595–1600. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Herron, M.D.; Howell, K.; Pentz, J.T.; Rosenzweig, F.; Travisano, M. Experimental evolution of an alternating uni-and multicellular life cycle in Chlamydomonas reinhardtii. Nat. Commun. 2013, 4, 2742. [Google Scholar] [CrossRef]
- Becks, L.; Ellner, S.P.; Jones, L.E.; Hairston, N.G., Jr. Reduction of adaptive genetic diversity radically alters eco-evolutionary community dynamics. Ecol. Lett. 2010, 13, 989–997. [Google Scholar] [CrossRef]
- Herron, M.D.; Borin, J.M.; Boswell, J.C.; Walker, J.; Chen, I.C.K.; Knox, C.A.; Boyd, M.; Rosenzweig, F.; Ratcliff, W.C. De novo origins of multicellularity in response to predation. Sci. Rep. 2019, 9, 2328. [Google Scholar] [CrossRef]
- Maynard Smith, J.; Szathmáry, E. The Major Transitions in Evolution; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Michod, R.E.; Roze, D. Cooperation and conflict in the evolution of multicellularity. Heredity 2001, 86, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Griesemer, J. The units of evolutionary transition. Selection 2001, 1, 67–80. [Google Scholar] [CrossRef]
- Okasha, S. Evolution and the Levels of Selection; Clarendon Press: Oxford, UK, 2006. [Google Scholar]
- Rainey, P.B.; Kerr, B. Cheats as first propagules: A new hypothesis for the evolution of individuality during the transition from single cells to multicellularity. BioEssays 2010, 32, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Trestman, M. Which comes first in major transitions: The behavioral chicken, or the evolutionary Egg? Biol. Theory 2013, 7, 48–55. [Google Scholar] [CrossRef]
- Simpson, C. How many levels are there? How insights from evolutionary transitions in individuality help measure the hierarchical complexity of life. In The Major Transitions in Evolution Revisited; Calcott, B., Sterelny, K., Eds.; The MIT Press: Cambridge, MA, USA, 2011; Chapter 10; pp. 199–226. [Google Scholar]
- Müller, G.; Newman, S. Generation, integration, autonomy: Three steps in the evolution of homology. In Proceedings of the Novartis Foundation Symposium, London, UK, 2–4 March 1999; Volume 222, pp. 65–79. [Google Scholar]
- Newman, S.A.; Müller, G.B. Epigenetic mechanisms of character origination. J. Exp. Zool. 2000, 288, 304–317. [Google Scholar] [CrossRef]
- Alvarado, A.S.; Kang, H. Multicellularity, stem cells, and the neoblasts of the planarian Schmidtea mediterranea. Exp. Cell Res. 2005, 306, 299–308. [Google Scholar] [CrossRef]
- Libby, E.; Rainey, P.B. A conceptual framework for the evolutionary origins of multicellularity. Phys. Biol. 2013, 10, 035001. [Google Scholar] [CrossRef]
- Michod, R.E. Evolution of the individual. Am. Nat. 1997, 150, S5–S21. [Google Scholar] [CrossRef]
- Michod, R.E.; Nedelcu, A.M.; Roze, D. Cooperation and conflict in the evolution of individuality: IV. Conflict mediation and evolvability in Volvox carteri. BioSystems 2003, 69, 95–114. [Google Scholar] [CrossRef]
- Michod, R.E.; Nedelcu, A.M. Cooperation and conflict during the unicellular-multicellular and prokaryotic-eukaryotic transitions. In Evolution: From Molecules to Ecosystems; Oxford University Press: Oxford, UK, 2003; pp. 195–208. [Google Scholar]
- Nedelcu, A.M.; Michod, R.E. Evolvability, modularity, and individuality during the transition to multicellularity in volvocalean green algae. In Modularity in Development and Evolution; Wagner, G., Schlosser, G., Eds.; University of Chicago Press: Chicago, IL, USA, 2004; pp. 468–489. [Google Scholar]
- Rainey, P.B.; De Monte, S. Resolving conflicts during the evolutionary transition to multicellular life. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 599–620. [Google Scholar] [CrossRef]
- Michod, R.E. Multi-Level Selection of the Individual Organism. In The Evolution of Multicellularity; CRC Press: Boca Raton, FL, USA, 2022; pp. 25–52. [Google Scholar]
- Herron, M.D.; Zamani-Dahaj, S.A.; Ratcliff, W.C. Trait heritability in major transitions. BMC Biol. 2018, 16, 145. [Google Scholar] [CrossRef] [PubMed]
- Bozdag, G.O.; Libby, E.; Pineau, R.; Reinhard, C.T.; Ratcliff, W.C. Oxygen suppression of macroscopic multicellularity. Nat. Commun. 2021, 12, 2838. [Google Scholar] [CrossRef] [PubMed]
- Bozdag, G.O.; Zamani-Dahaj, S.A.; Day, T.C.; Kahn, P.C.; Burnetti, A.J.; Lac, D.T.; Tong, K.; Conlin, P.L.; Balwani, A.H.; Dyer, E.L.; et al. De novo evolution of macroscopic multicellularity. Nature 2023, 617, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Jacobeen, S.; Pentz, J.T.; Graba, E.C.; Brandys, C.G.; Ratcliff, W.C.; Yunker, P.J. Cellular packing, mechanical stress and the evolution of multicellularity. Nat. Phys. 2018, 14, 286–290. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer: Sunderland, MA, USA, 1998; Volume 1. [Google Scholar]
- Jacobeen, S.; Graba, E.C.; Brandys, C.G.; Day, T.C.; Ratcliff, W.C.; Yunker, P.J. Geometry, packing, and evolutionary paths to increased multicellular size. Phys. Rev. E 2018, 97, 050401. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Fankhauser, J.D.; Rogers, D.W.; Greig, D.; Travisano, M. Origins of multicellular evolvability in snowflake yeast. Nat. Commun. 2015, 6, 6102. [Google Scholar] [CrossRef]
- Jin, M.; Li, Y.; O’Laughlin, R.; Bittihn, P.; Pillus, L.; Tsimring, L.S.; Hasty, J.; Hao, N. Divergent aging of isogenic yeast cells revealed through single-cell phenotypic dynamics. Cell Syst. 2019, 8, 242–253. [Google Scholar] [CrossRef]
- Fisher, R.A. XV.—The correlation between relatives on the supposition of Mendelian inheritance. Earth Environ. Sci. Trans. R. Soc. Edinb. 1919, 52, 399–433. [Google Scholar] [CrossRef]
- Herron, M.D. What are the major transitions? Biol. Philos. 2021, 36, 2. [Google Scholar] [CrossRef]
- Lewontin, R.C. The units of selection. Annu. Rev. Ecol. Syst. 1970, 1, 1–18. [Google Scholar] [CrossRef]
- Schaap, P.; Winckler, T.; Nelson, M.; Alvarez-Curto, E.; Elgie, B.; Hagiwara, H.; Cavender, J.; Milano-Curto, A.; Rozen, D.E.; Dingermann, T.; et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science 2006, 314, 661–663. [Google Scholar] [CrossRef]
- Schaap, P. Eukaryote Aggregative Multicellularity: Phylogenetic Distribution and a Case Study of Its Proximate and Ultimate Cause in Dictyostelia. In The Evolution of Multicellularity; CRC Press: Boca Raton, FL, USA, 2022; pp. 73–88. [Google Scholar]
- Grosberg, R.K.; Strathmann, R.R. The evolution of multicellularity: A minor major transition? Annu. Rev. Ecol. Evol. Syst. 2007, 38, 621–654. [Google Scholar] [CrossRef]
- Umen, J.; Herron, M.D. Green algal models for multicellularity. Annu. Rev. Genet. 2021, 55, 603–632. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Conlin, P.L.; Herron, M.D. Conclusion: The Future of Multicellularity Research. In The Evolution of Multicellularity; CRC Press: Boca Raton, FL, USA, 2022; pp. 359–372. [Google Scholar]
- Lamża, Ł. Diversity of ‘simple’ multicellular eukaryotes: 45 independent cases and six types of multicellularity. Biol. Rev. 2023. [Google Scholar] [CrossRef] [PubMed]
- Bourrat, P. Evolutionary transitions in heritability and individuality. Theory Biosci. 2019, 138, 305–323. [Google Scholar] [CrossRef] [PubMed]
- Price, G.R. Selection and covariance. Nature 1970, 227, 520–521. [Google Scholar] [CrossRef] [PubMed]
- Queller, D.C. Quantitative genetics, inclusive fitness, and group selection. Am. Nat. 1992, 139, 540–558. [Google Scholar] [CrossRef]
- Gardner, A. The genetical theory of multilevel selection. J. Evol. Biol. 2015, 28, 305–319. [Google Scholar] [CrossRef]
- Goodnight, C.J. Multilevel selection theory and evidence: A critique of Gardner, 2015. J. Evol. Biol. 2015, 28, 1734–1746. [Google Scholar] [CrossRef]
- Burton, G.W.; Devane, D.E. Estimating heritability in tall fescue (Festuca arundinacea) from replicated clonal material 1. Agron. J. 1953, 45, 478–481. [Google Scholar] [CrossRef]
- Bokmeyer, J.M.; Bonos, S.A.; Meyer, W.A. Broad-sense heritability and stability analysis of brown patch resistance in tall fescue. HortScience 2009, 44, 289–292. [Google Scholar] [CrossRef]
- Bonos, S.A.; Casler, M.D.; Meyer, W.A. Inheritance of dollar spot resistance in creeping bentgrass. Crop. Sci. 2003, 43, 2189–2196. [Google Scholar] [CrossRef]
- Stratton, D.A. Life history variation within populations of an asexual plant, Erigeron annuus (Asteraceae). Am. J. Bot. 1991, 78, 723–728. [Google Scholar] [CrossRef]
- Oard, J.; Simons, M. A growth chamber comparison of traits of aggressiveness in sexual and asexual populations of Puccinia coronata. Phytopathology 1983, 73, 1226–1229. [Google Scholar] [CrossRef]
- Weisser, W.W.; Braendle, C. Body colour and genetic variation in winged morph production in the pea aphid. Entomol. Exp. Appl. 2001, 99, 217–223. [Google Scholar] [CrossRef]
- Vorburger, C. Positive genetic correlations among major life-history traits related to ecological success in the pea aphid Myzus perisicae. Evolution 2005, 59, 1006–1015. [Google Scholar] [PubMed]
- De Meester, L. An estimation of the heritability of phototaxis in Daphnia magna Straus. Oecologia 1989, 78, 142–144. [Google Scholar] [CrossRef]
- Yampolsky, L.Y.; Ebert, D. Variation and plasticity of biomass allocation in Daphnia. Funct. Ecol. 1994, 8, 435–440. [Google Scholar] [CrossRef]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef]
- Dykhuizen, D.E. Experimental studies of natural selection in bacteria. Annu. Rev. Ecol. Syst. 1990, 21, 373–398. [Google Scholar] [CrossRef]
- Bouma, J.E.; Lenski, R.E. Evolution of a bacteria/plasmid association. Nature 1988, 335, 351–352. [Google Scholar] [CrossRef]
- López Rodas, V.; Maneiro, E.; Costas, E. Adaptation of cyanobacteria and microalgae to extreme environmental changes derived from anthropogenic pollution. Limnetica 2006, 25, 403–410. [Google Scholar] [CrossRef]
- Baselga-Cervera, B.; Costas, E.; Bustillo-Avendaño, E.; García-Balboa, C. Adaptation prevents the extinction of Chlamydomonas reinhardtii under toxic beryllium. PeerJ 2016, 4, e1823. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, A.; Cleathero, L.; Mandegar, M.; Otto, S. Haploids adapt faster than diploids across a range of environments. J. Evol. Biol. 2011, 24, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Vogt, G. Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences. J. Biosci. 2015, 40, 159–204. [Google Scholar] [CrossRef] [PubMed]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic Gene Expression in a Single Cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef]
- Ballouz, S.; Pena, M.T.; Knight, F.M.; Adams, L.B.; Gillis, J.A. The Transcriptional Legacy of Developmental Stochasticity. bioRxiv 2019. Available online: http://xxx.lanl.gov/abs/2019.12.11.873265 (accessed on 26 July 2023). [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Lee, J.A.; Riazi, S.; Nemati, S.; Bazurto, J.V.; Vasdekis, A.E.; Ridenhour, B.J.; Remien, C.H.; Marx, C.J. Microbial phenotypic heterogeneity in response to a metabolic toxin: Continuous, dynamically shifting distribution of formaldehyde tolerance in Methylobacterium extorquens populations. PLos Genet. 2019, 15, e1008458. [Google Scholar] [CrossRef]
- Salek, M.M.; Carrara, F.; Fernandez, V.; Guasto, J.S.; Stocker, R. Bacterial chemotaxis in a microfluidic T-maze reveals strong phenotypic heterogeneity in chemotactic sensitivity. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Kain, J.S.; Stokes, C.; de Bivort, B.L. Phototactic personality in fruit flies and its suppression by serotonin and white. Proc. Natl. Acad. Sci. USA 2012, 109, 19834–19839. [Google Scholar] [CrossRef] [PubMed]
- Herron, M.D.; Ghimire, S.; Vinikoor, C.R.; Michod, R.E. Fitness trade-offs and developmental constraints in the evolution of soma: An experimental study in a volvocine alga. Evol. Ecol. Res. 2014, 16, 203. [Google Scholar]
- Schuett, W.; Dall, S.R.X.; Baeumer, J.; Kloesener, M.H.; Nakagawa, S.; Beinlich, F.; Eggers, T. Personality variation in a clonal insect: The pea aphid, Acyrthosiphon pisum. Dev. Psychobiol. 2011, 53, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Vogt, G.; Huber, M.; Thiemann, M.; van den Boogaart, G.; Schmitz, O.J.; Schubart, C.D. Production of different phenotypes from the same genotype in the same environment by developmental variation. J. Exp. Biol. 2008, 211, 510–523. [Google Scholar] [CrossRef]
- Okasha, S. The concept of group heritability. Biol. Philos. 2003, 18, 445–461. [Google Scholar] [CrossRef]
- Okasha, S. Multilevel selection and the major transitions in evolution. Philos. Sci. 2013, 72, 1013–1025. [Google Scholar] [CrossRef]
- Doulcier, G.; Lambert, A.; Monte, S.D.; Rainey, P.B. Eco-evolutionary dynamics of nested Darwinian populations and the emergence of community-level heredity. eLife 2020, 9, e53433. [Google Scholar] [CrossRef]
- Clarke, E. Origins of evolutionary transitions. J. Biosci. 2014, 39, 303–317. [Google Scholar] [CrossRef]
- Libby, E.; Ratcliff, W.C. Ratcheting the evolution of multicellularity. Science 2014, 346, 426–427. [Google Scholar] [CrossRef]
- Michod, R.E. Darwinian Dynamics: Evolutionary Transitions in Fitness and Individuality; Princeton University Press: Princeton, NJ, USA, 1999. [Google Scholar]
- Rainey, P.B.; Kerr, B. Conflicts among levels of selection as fuel for the evolution of individuality. Major Transi. Evol. Revisit. 2011, 22, 141–162. [Google Scholar]
- Edgington, N.P.; Blacketer, M.J.; Bierwagen, T.A.; Myers, A.M. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol. Cell. Biol. 1999, 19, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Watanabe, D.; Nogami, S.; Morishita, S.; Ohya, Y. Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr. Genet. 2009, 55, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Woods, R.A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. In Yeast Protocol; Springer: Berlin/Heidelberg, Germany, 2006; pp. 107–120. [Google Scholar]
- Janke, C.; Magiera, M.M.; Rathfelder, N.; Taxis, C.; Reber, S.; Maekawa, H.; Moreno-Borchart, A.; Doenges, G.; Schwob, E.; Schiebel, E.; et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 2004, 21, 947–962. [Google Scholar] [CrossRef]
- Bourrat, P. Unifying heritability in evolutionary theory. Stud. Hist. Philos. Sci. 2022, 91, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Mameli, M. Nongenetic selection and nongenetic inheritance. Br. J. Philos. Sci. 2004, 55, 31–75. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zamani-Dahaj, S.A.; Burnetti, A.; Day, T.C.; Yunker, P.J.; Ratcliff, W.C.; Herron, M.D. Spontaneous Emergence of Multicellular Heritability. Genes 2023, 14, 1635. https://doi.org/10.3390/genes14081635
Zamani-Dahaj SA, Burnetti A, Day TC, Yunker PJ, Ratcliff WC, Herron MD. Spontaneous Emergence of Multicellular Heritability. Genes. 2023; 14(8):1635. https://doi.org/10.3390/genes14081635
Chicago/Turabian StyleZamani-Dahaj, Seyed Alireza, Anthony Burnetti, Thomas C. Day, Peter J. Yunker, William C. Ratcliff, and Matthew D. Herron. 2023. "Spontaneous Emergence of Multicellular Heritability" Genes 14, no. 8: 1635. https://doi.org/10.3390/genes14081635
APA StyleZamani-Dahaj, S. A., Burnetti, A., Day, T. C., Yunker, P. J., Ratcliff, W. C., & Herron, M. D. (2023). Spontaneous Emergence of Multicellular Heritability. Genes, 14(8), 1635. https://doi.org/10.3390/genes14081635





