Enhancer Function in the 3D Genome
Abstract
1. Introduction
2. The Mechanism of Enhancers Action
3. Establishing Communication between Enhancers and Promoters
4. Chromatin Hubs or Chromatin Compartments
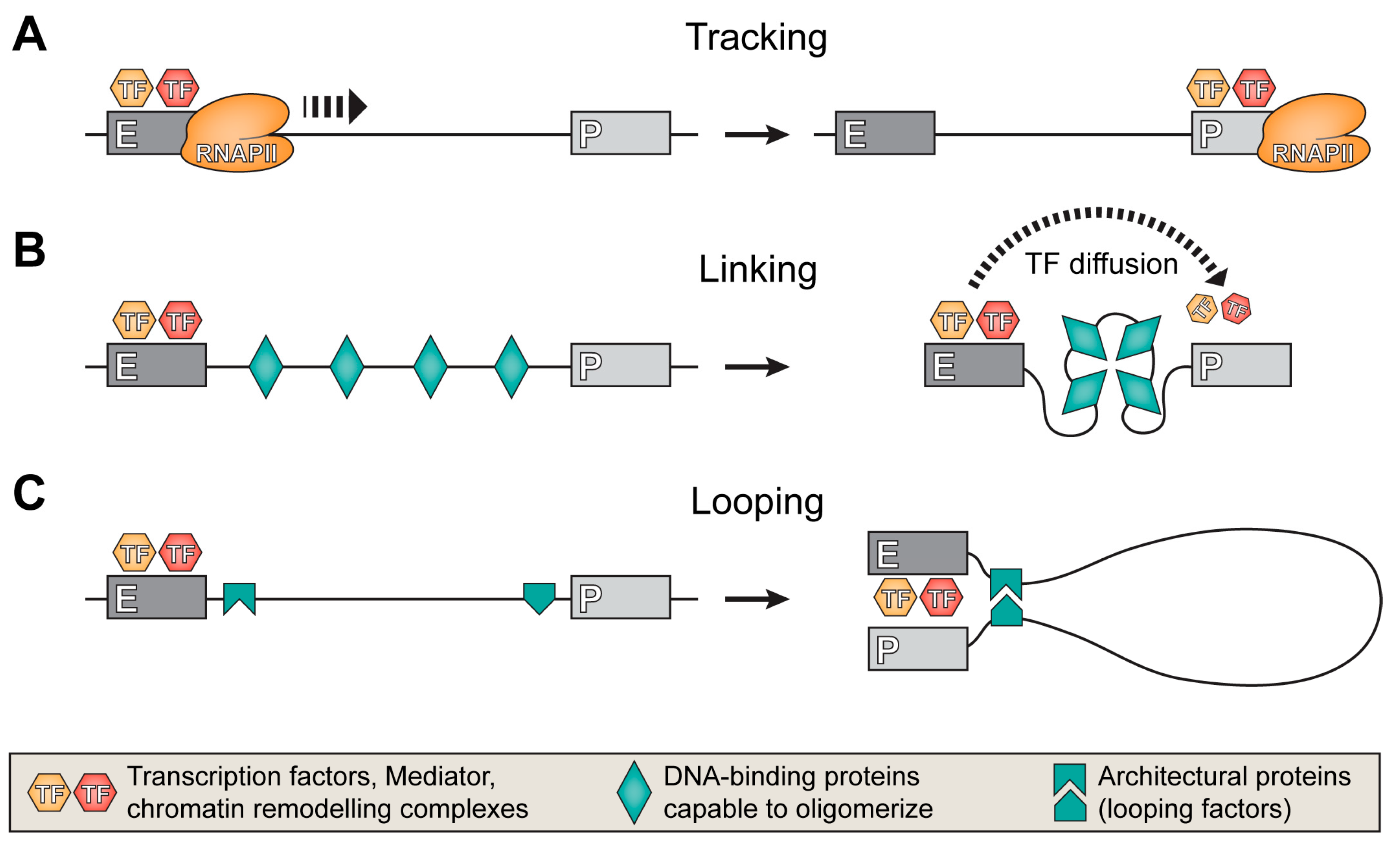
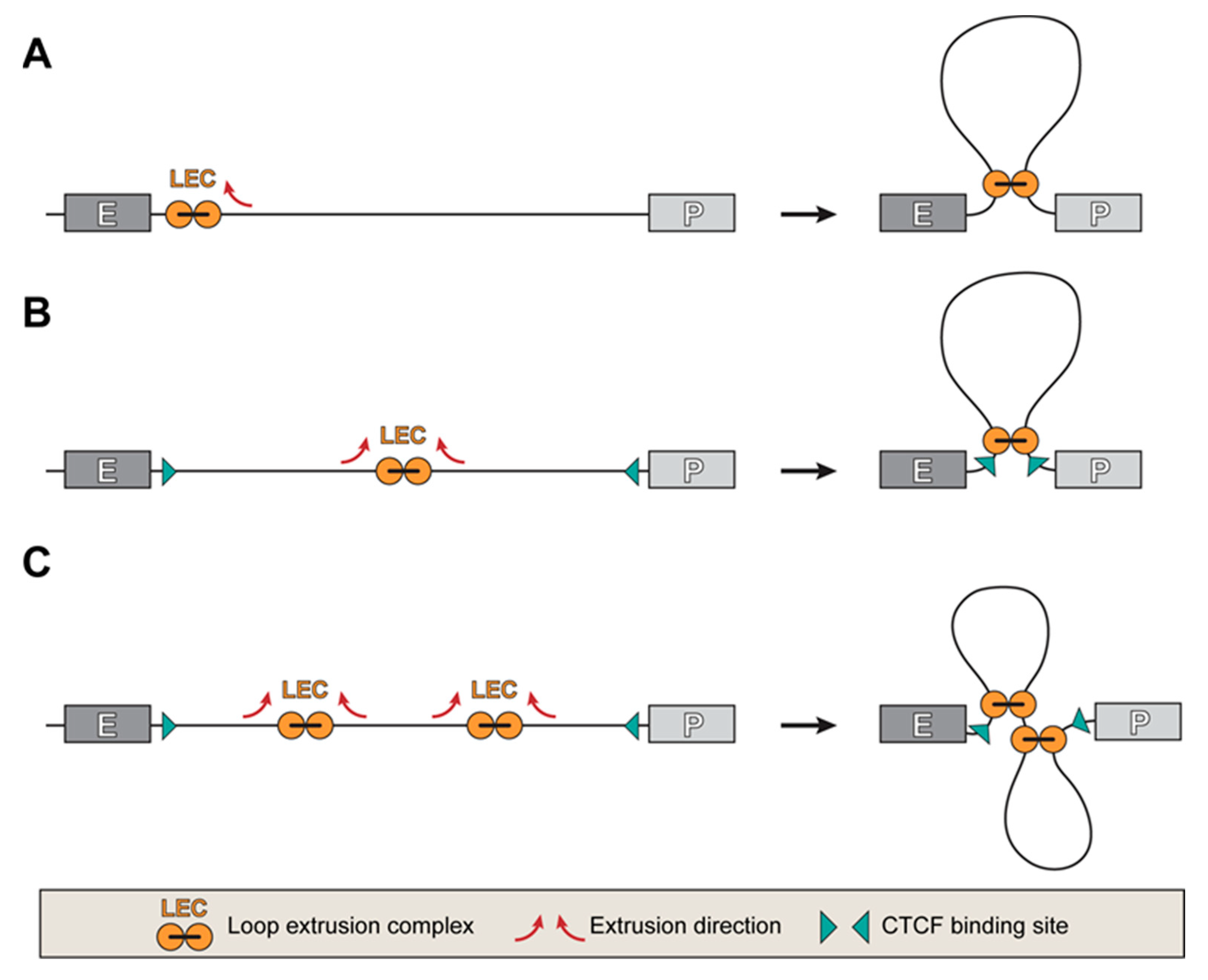
5. Mechanisms of the Target Promoter Search
6. Selective Activation of Certain Promoters by Remote Enhancers
7. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Banerji, J.; Rusconi, S.; Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308. [Google Scholar] [CrossRef]
- Moreau, P.; Hen, R.; Wasylyk, B.; Everett, R.; Gaub, M.P.; Chambon, P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981, 9, 6047–6068. [Google Scholar] [CrossRef]
- Dorsch-Häsler, K.; Keil, G.M.; Weber, F.; Jasin, M.; Schaffner, W.; Koszinowski, U.H. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc. Natl. Acad. Sci. USA 1985, 82, 8325–8329. [Google Scholar] [CrossRef]
- de Villiers, J.; Schaffner, W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981, 9, 6251–6264. [Google Scholar] [CrossRef]
- de Villiers, J.; Olson, L.; Banerji, J.; Schaffner, W. Analysis of the Transcriptional Enhancer Effect. Cold Spring Harb. Symp. Quant. Biol. 1983, 47 Pt 2, 911–919. [Google Scholar] [CrossRef]
- Muller, M.M.; Gerster, T.; Schaffner, W. Enhancer sequences and the regulation of gene transcription. Eur. J. Biochem. 1988, 176, 485–495. [Google Scholar] [CrossRef]
- Schaffner, W. Enhancers, enhancers—From their discovery to today’s universe of transcription enhancers. Biol. Chem. 2015, 396, 311–327. [Google Scholar] [CrossRef]
- Maniatis, T.; Goodbourn, S.; Fischer, J.A. Regulation of Inducible and Tissue-Specific Gene Expression. Science 1987, 236, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.V.; Bich-Thuy, L.T.; Stafford, J.; Queen, C. Synergism between immunoglobulin enhancers and promoters. Nature 1986, 322, 383–385. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Carey, M. The Enhanceosome and Transcriptional Synergy. Cell 1998, 92, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Panne, D. The enhanceosome. Curr. Opin. Struct. Biol. 2008, 18, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Arnosti, D.N.; Kulkarni, M.M. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 2005, 94, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Grosveld, F.; van Staalduinen, J.; Stadhouders, R. Transcriptional Regulation by (Super)Enhancers: From Discovery to Mechanisms. Annu. Rev. Genom. Hum. Genet. 2021, 22, 127–146. [Google Scholar] [CrossRef]
- Talbot, D.; Collis, P.; Antoniou, M.; Vidal, M.; Grosveld, F.; Greaves, D.R. A dominant control region from the human b-globin locus conferring integration site-independent gene expression. Nature 1989, 338, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Peterson, K.R.; Fang, X.; Stamatoyannopoulos, G. Locus control regions. Blood 2002, 100, 3077–3086. [Google Scholar] [CrossRef]
- Pott, S.; Lieb, J.D. What are super-enhancers? Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cairns, M.J.; Yan, J. Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 2019, 47, 11481–11496. [Google Scholar] [CrossRef]
- Heinz, S.; Romanoski, C.E.; Benner, C.; Glass, C.K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015, 16, 144–154. [Google Scholar] [CrossRef]
- Lewis, M.W.; Li, S.; Franco, H.L. Transcriptional control by enhancers and enhancer RNAs. Transcription 2019, 10, 171–186. [Google Scholar] [CrossRef]
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef]
- Consortium, E.P.; Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Thurman, R.E.; Rynes, E.; Humbert, R.; Vierstra, J.; Maurano, M.T.; Haugen, E.; Sheffield, N.C.; Stergachis, A.B.; Wang, H.; Vernot, B.; et al. The accessible chromatin landscape of the human genome. Nature 2012, 489, 75–82. [Google Scholar] [CrossRef]
- Parsi, K.M.; Hennessy, E.; Kearns, N.; Maehr, R. Using an Inducible CRISPR-dCas9-KRAB Effector System to Dissect Transcriptional Regulation in Human Embryonic Stem Cells. Methods Mol. Biol. 2017, 1507, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, M.; Hill, A.J.; McFaline-Figueroa, J.L.; Martin, B.; Kim, S.; Zhang, M.D.; Jackson, D.; Leith, A.; Schreiber, J.; Noble, W.S.; et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 2019, 176, 377–390.e319. [Google Scholar] [CrossRef] [PubMed]
- Patrinos, G.P.; de Krom, M.; de Boer, E.; Langeveld, A.; Imam, A.M.A.; Strouboulis, J.; de Laat, W.; Grosveld, F.G. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004, 18, 1495–1509. [Google Scholar] [CrossRef] [PubMed]
- Farooq, U.; Saravanan, B.; Islam, Z.; Walavalkar, K.; Singh, A.K.; Jayani, R.S.; Meel, S.; Swaminathan, S.; Notani, D. An interdependent network of functional enhancers regulates transcription and EZH2 loading at the INK4a/ARF locus. Cell Rep. 2021, 34, 108898. [Google Scholar] [CrossRef]
- Cullen, K.E.; Kladde, M.P.; Seyfred, M.A. Interaction between Transcription Regulatory Regions of Prolactin Chromatin. Science 1993, 261, 203–206. [Google Scholar] [CrossRef]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing Chromosome Conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- de Wit, E.; de Laat, W. A decade of 3C technologies: Insights into nuclear organization. Genes Dev. 2012, 26, 11–24. [Google Scholar] [CrossRef]
- Denker, A.; de Laat, W. The second decade of 3C technologies: Detailed insights into nuclear organization. Genes Dev. 2016, 30, 1357–1382. [Google Scholar] [CrossRef]
- Tolhuis, B.; Palstra, R.-J.; Splinter, E.; Grosveld, F.; de Laat, W. Looping and Interaction between Hypersensitive Sites in the Active beta-globin Locus. Mol. Cell 2002, 10, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Vernimmen, D.; De Gobbi, M.; A Sloane-Stanley, J.A.; Wood, W.G.; Higgs, D.R. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007, 26, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Williamson, I.; Lettice, L.A.; Hill, R.E.; Bickmore, W.A. Shh and ZRS enhancer co-localisation is specific to the zone of polarizing activity. Development 2016, 143, 2994–3001. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef]
- Symmons, O.; Uslu, V.V.; Tsujimura, T.; Ruf, S.; Nassari, S.; Schwarzer, W.; Ettwiller, L.; Spitz, F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014, 24, 390–400. [Google Scholar] [CrossRef]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef]
- Lupiáñez, D.G.; Spielmann, M.; Mundlos, S. Breaking TADs: How Alterations of Chromatin Domains Result in Disease. Trends Genet. 2016, 32, 225–237. [Google Scholar] [CrossRef]
- Kaiser, V.B.; Semple, C.A. When TADs go bad: Chromatin structure and nuclear organisation in human disease. F1000Research 2017, 6, 314. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Sanborn, A.L.; Rao, S.S.P.; Huang, S.-C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.P.; Huang, S.-C.; St Hilaire, B.G.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e324. [Google Scholar] [CrossRef]
- Kim, Y.; Shi, Z.; Zhang, H.; Finkelstein, I.J.; Yu, H. Human cohesin compacts DNA by loop extrusion. Science 2019, 366, 1345–1349. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e922. [Google Scholar] [CrossRef]
- Wutz, G.; Várnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef]
- Davidson, I.F.; Barth, R.; Zaczek, M.; van der Torre, J.; Tang, W.; Nagasaka, K.; Janissen, R.; Kerssemakers, J.; Wutz, G.; Dekker, C.; et al. CTCF is a DNA-tension-dependent barrier to cohesin-mediated loop extrusion. Nature 2023, 616, 822–827. [Google Scholar] [CrossRef]
- Chubb, J.R.; Liverpool, T.B. Bursts and pulses: Insights from single cell studies into transcriptional mechanisms. Curr. Opin. Genet. Dev. 2010, 20, 478–484. [Google Scholar] [CrossRef]
- Chubb, J.R.; Trcek, T.; Shenoy, S.M.; Singer, R.H. Transcriptional Pulsing of a Developmental Gene. Curr. Biol. 2006, 16, 1018–1025. [Google Scholar] [CrossRef]
- Rodriguez, J.; Larson, D.R. Transcription in Living Cells: Molecular Mechanisms of Bursting. Annu. Rev. Biochem. 2020, 89, 189–212. [Google Scholar] [CrossRef]
- Fukaya, T.; Lim, B.; Levine, M. Enhancer Control of Transcriptional Bursting. Cell 2016, 166, 358–368. [Google Scholar] [CrossRef]
- Bartman, C.R.; Hsu, S.C.; Hsiung, C.C.-S.; Raj, A.; Blobel, G.A. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol. Cell 2016, 62, 237–247. [Google Scholar] [CrossRef]
- Cheng, L.; De, C.; Li, J.; Pertsinidis, A. Mechanisms of transcription control by distal enhancers from high-resolution single-gene imaging. bioRxiv 2023, 3, 233190. [Google Scholar] [CrossRef]
- Larsson, A.J.M.; Johnsson, P.; Hagemann-Jensen, M.; Hartmanis, L.; Faridani, O.R.; Reinius, B.; Segerstolpe, Å.; Rivera, C.M.; Ren, B.; Sandberg, R. Genomic encoding of transcriptional burst kinetics. Nature 2019, 565, 251–254. [Google Scholar] [CrossRef]
- De Santa, F.; Barozzi, I.; Mietton, F.; Ghisletti, S.; Polletti, S.; Tusi, B.K.; Muller, H.; Ragoussis, J.; Wei, C.-L.; Natoli, G. A Large Fraction of Extragenic RNA Pol II Transcription Sites Overlap Enhancers. PLoS Biol. 2010, 8, e1000384. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Du, G.; Song, X.; Li, L. Non-coding Transcripts from Enhancers: New Insights into Enhancer Activity and Gene Expression Regulation. Genom. Proteom. Bioinform. 2017, 15, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Kaikkonen, M.U.; Spann, N.J.; Heinz, S.; Romanoski, C.E.; Allison, K.A.; Stender, J.D.; Chun, H.B.; Tough, D.F.; Prinjha, R.K.; Benner, C.; et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Mol. Cell 2013, 51, 310–325. [Google Scholar] [CrossRef] [PubMed]
- Mikhaylichenko, O.; Bondarenko, V.; Harnett, D.; Schor, I.E.; Males, M.; Viales, R.R.; Furlong, E.E.M. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 2018, 32, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.R.; Wells, A.D.; Li, X.C. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front. Cell Dev. Biol. 2020, 7, 377. [Google Scholar] [CrossRef]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef]
- De Lara, J.C.-F.; Arzate-Mejía, R.G.; Recillas-Targa, F. Enhancer RNAs: Insights into Their Biological Role. Epigenetics Insights 2019, 12, 2516865719846093. [Google Scholar] [CrossRef]
- Sigova, A.A.; Abraham, B.J.; Ji, X.; Molinie, B.; Hannett, N.M.; Guo, Y.E.; Jangi, M.; Giallourakis, C.C.; Sharp, P.A.; Young, R.A. Transcription factor trapping by RNA in gene regulatory elements. Science 2015, 350, 978–981. [Google Scholar] [CrossRef]
- Razin, S.V.; Gavrilov, A.A. Non-coding RNAs in chromatin folding and nuclear organization. Cell. Mol. Life Sci. 2021, 78, 5489–5504. [Google Scholar] [CrossRef]
- Lee, J.-H.; Wang, R.; Xiong, F.; Krakowiak, J.; Liao, Z.; Nguyen, P.T.; Moroz-Omori, E.V.; Shao, J.; Zhu, X.; Bolt, M.J.; et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 2021, 81, 3368–3385.e3369. [Google Scholar] [CrossRef]
- Hsieh, C.-L.; Fei, T.; Chen, Y.; Li, T.; Gao, Y.; Wang, X.; Sun, T.; Sweeney, C.J.; Lee, G.-S.M.; Chen, S.; et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. USA 2014, 111, 7319–7324. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Andersson, R. Promoter or enhancer, what’s the difference? Deconstruction of established distinctions and presentation of a unifying model. Bioessays 2015, 37, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef]
- Dao, L.T.M.; O Galindo-Albarrán, A.; Castro-Mondragon, J.A.; Andrieu-Soler, C.; Rivera, A.M.; Souaid, C.; Charbonnier, G.; Griffon, A.; Vanhille, L.; Stephen, T.; et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 2017, 49, 1073–1081. [Google Scholar] [CrossRef]
- Dao, L.T.M.; Spicuglia, S. Transcriptional regulation by promoters with enhancer function. Transcription 2018, 9, 307–314. [Google Scholar] [CrossRef]
- Medina-Rivera, A.; Santiago-Algarra, D.; Puthier, D.; Spicuglia, S. Widespread Enhancer Activity from Core Promoters. Trends Biochem. Sci. 2018, 43, 452–468. [Google Scholar] [CrossRef] [PubMed]
- Zabidi, M.A.; Arnold, C.D.; Schernhuber, K.; Pagani, M.; Rath, M.; Frank, O.; Stark, A. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature 2015, 518, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, M.S.; Hughes, J.R.; Garrick, D.; Lynch, M.D.; Sharpe, J.A.; Sloane-Stanley, J.A.; McGowan, S.J.; De Gobbi, M.; Hosseini, M.; Vernimmen, D.; et al. Intragenic Enhancers Act as Alternative Promoters. Mol. Cell 2012, 45, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Travers, A. Chromatin modification by DNA tracking. Proc. Natl. Acad. Sci. USA 1999, 96, 13634–13637. [Google Scholar] [CrossRef]
- Dorsett, D. Distant liaisons: Long-range enhancer–promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 1999, 9, 505–514. [Google Scholar] [CrossRef]
- Bulger, M.; Groudine, M. Looping versus linking: Toward a model for long-distance gene activation. Genes Dev. 1999, 13, 2465–2477. [Google Scholar] [CrossRef]
- Ptashne, M. Gene regulation by proteins acting nearby and at a distance. Nature 1986, 322, 697–701. [Google Scholar] [CrossRef]
- Jin, F.; Li, Y.; Dixon, J.R.; Selvaraj, S.; Ye, Z.; Lee, A.Y.; Yen, C.-A.; Schmitt, A.D.; Espinoza, C.A.; Ren, B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 2013, 503, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Vernimmen, D.; Marques-Kranc, F.; Sharpe, J.A.; Sloane-Stanley, J.A.; Wood, W.G.; Wallace, H.A.C.; Smith, A.J.H.; Higgs, D.R. Chromosome looping at the human alpha-globin locus is mediated via the major upstream regulatory element (HS−40). Blood 2009, 114, 4253–4260. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Gavrilov, A.A.; Ioudinkova, E.S.; Iarovaia, O.V. Communication of genome regulatory elements in a folded chromosome. FEBS Lett. 2013, 587, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Kyrchanova, O.V.; Bylino, O.V.; Georgiev, P.G. Mechanisms of enhancer-promoter communication and chromosomal architecture in mammals and Drosophila. Front. Genet. 2022, 13, 1081088. [Google Scholar] [CrossRef] [PubMed]
- Popay, T.M.; Dixon, J.R. Coming full circle: On the origin and evolution of the looping model for enhancer–promoter communication. J. Biol. Chem. 2022, 298, 102117. [Google Scholar] [CrossRef]
- Palstra, R.-J.T.S. Close encounters of the 3C kind: Long-range chromatin interactions and transcriptional regulation. Brief. Funct. Genom. Proteom. 2009, 8, 297–309. [Google Scholar] [CrossRef]
- Breda, L.; Motta, I.; Lourenco, S.; Gemmo, C.; Deng, W.; Rupon, J.W.; Abdulmalik, O.Y.; Manwani, D.; Blobel, G.A.; Rivella, S. Forced chromatin looping raises fetal hemoglobin in adult sickle cells to higher levels than pharmacologic inducers. Blood 2016, 128, 1139–1143. [Google Scholar] [CrossRef]
- Deng, W.; Lee, J.; Wang, H.; Miller, J.; Reik, A.; Gregory, P.D.; Dean, A.; Blobel, G.A. Controlling Long-Range Genomic Interactions at a Native Locus by Targeted Tethering of a Looping Factor. Cell 2012, 149, 1233–1244. [Google Scholar] [CrossRef]
- Benabdallah, N.S.; Williamson, I.; Illingworth, R.S.; Kane, L.; Boyle, S.; Sengupta, D.; Grimes, G.R.; Therizols, P.; Bickmore, W.A. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 2019, 76, 473–484.e477. [Google Scholar] [CrossRef]
- Golov, A.K.; Gavrilov, A.A.; Kaplan, N.; Razin, S.V. A genome-wide nucleosome-resolution map of promoter-centered interactions in human cells corroborates the enhancer-promoter looping model. bioRxiv 2023, 13, 528105. [Google Scholar] [CrossRef]
- Maksimenko, O.; Golovnin, A.; Georgiev, P. Enhancer-Promoter Communication Is Regulated by Insulator Pairing in a Drosophila Model Bigenic Locus. Mol. Cell. Biol. 2008, 28, 5469–5477. [Google Scholar] [CrossRef]
- Savitskaya, E.; Melnikova, L.; Kostuchenko, M.; Kravchenko, E.; Pomerantseva, E.; Boikova, T.; Chetverina, D.; Parshikov, A.; Zobacheva, P.; Gracheva, E.; et al. Study of Long-Distance Functional Interactions between Su(Hw) Insulators That Can Regulate Enhancer-Promoter Communication in Drosophila melanogaster. Mol. Cell. Biol. 2006, 26, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Peters, J.-M. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 445–464. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.J.; Aguilar, R.; Kesner, B.; Lee, H.-G.; Kriz, A.J.; Chu, H.-P.; Lee, J.T. Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell 2021, 184, 6157–6173.e6124. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Galitsyna, A.A.; Flyamer, I.M.; Golov, A.K.; Khrameeva, E.E.; Imakaev, M.V.; Abdennur, N.A.; Gelfand, M.S.; Gavrilov, A.A.; Razin, S.V. Activation of the α-globin gene expression correlates with dramatic upregulation of nearby non-globin genes and changes in local and large-scale chromatin spatial structure. Epigenetics Chromatin 2017, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, L.L.P.; Kassouf, M.T.; Oudelaar, A.M.; Biggs, D.; Preece, C.; Downes, D.J.; Gosden, M.; Sharpe, J.A.; Sloane-Stanley, J.A.; Hughes, J.R.; et al. Tissue-specific CTCF–cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 2017, 19, 952–961. [Google Scholar] [CrossRef]
- Li, W.; Jiang, H. Nuclear Protein Condensates and Their Properties in Regulation of Gene Expression. J. Mol. Biol. 2022, 434, 167151. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef]
- Kantidze, O.L.; Razin, S.V. Weak interactions in higher-order chromatin organization. Nucleic Acids Res. 2020, 48, 4614–4626. [Google Scholar] [CrossRef]
- Gavrilov, A.A.; Gushchanskaya, E.S.; Strelkova, O.; Zhironkina, O.; Kireev, I.I.; Iarovaia, O.V.; Razin, S.V. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res. 2013, 41, 3563–3575. [Google Scholar] [CrossRef]
- Heist, T.; Fukaya, T.; Levine, M. Large distances separate coregulated genes in living Drosophila embryos. Proc. Natl. Acad. Sci. USA 2019, 116, 15062–15067. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Levo, M.; Barinov, L.; Fujioka, M.; Jaynes, J.B.; Gregor, T. Dynamic interplay between enhancer–promoter topology and gene activity. Nat. Genet. 2018, 50, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hsu, A.; Hua, Y.; Wang, G.; Cheng, L.; Ochiai, H.; Yamamoto, T.; Pertsinidis, A. Single-gene imaging links genome topology, promoter–enhancer communication and transcription control. Nat. Struct. Mol. Biol. 2020, 27, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.M.; Guan, J.; Li, B.; Maliskova, L.; Song, M.; Shen, Y.; Huang, B.; Lomvardas, S.; Weiner, O.D. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. Elife 2019, 8, e41769. [Google Scholar] [CrossRef]
- Goel, V.Y.; Huseyin, M.K.; Hansen, A.S. Region Capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments. Nat. Genet. 2023, 55, 1048–1056. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Velichko, A.K.; Magnitov, M.D.; Luzhin, A.V.; Golov, A.K.; Ovsyannikova, N.; I Kireev, I.; Gavrikov, A.S.; Mishin, A.S.; Garaev, A.K.; et al. Suppression of liquid–liquid phase separation by 1,6-hexanediol partially compromises the 3D genome organization in living cells. Nucleic Acids Res. 2021, 49, 10524–10541. [Google Scholar] [CrossRef]
- Finn, E.H.; Pegoraro, G.; Brandão, H.B.; Valton, A.-L.; Oomen, M.E.; Dekker, J.; Mirny, L.; Misteli, T. Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell 2019, 176, 1502–1515.e1510. [Google Scholar] [CrossRef]
- Gavrilov, A.A.; Golov, A.K.; Razin, S.V. Actual Ligation Frequencies in the Chromosome Conformation Capture Procedure. PLoS ONE 2013, 8, e60403. [Google Scholar] [CrossRef]
- Gavrilov, A.A.; Chetverina, H.V.; Chermnykh, E.S.; Razin, S.V.; Chetverin, A.B. Quantitative analysis of genomic element interactions by molecular colony technique. Nucleic Acids Res. 2014, 42, e36. [Google Scholar] [CrossRef]
- Morgan, S.L.; Mariano, N.C.; Bermudez, A.; Arruda, N.L.; Wu, F.; Luo, Y.; Shankar, G.; Jia, L.; Chen, H.; Hu, J.-F.; et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 2017, 8, 15993. [Google Scholar] [CrossRef]
- Deng, W.; Rupon, J.W.; Krivega, I.; Breda, L.; Motta, I.; Jahn, K.S.; Reik, A.; Gregory, P.D.; Rivella, S.; Dean, A.; et al. Reactivation of Developmentally Silenced Globin Genes by Forced Chromatin Looping. Cell 2014, 158, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Mirny, L. The 3D Genome as Moderator of Chromosomal Communication. Cell 2016, 164, 1110–1121. [Google Scholar] [CrossRef]
- Zuin, J.; Roth, G.; Zhan, Y.; Cramard, J.; Redolfi, J.; Piskadlo, E.; Mach, P.; Kryzhanovska, M.; Tihanyi, G.; Kohler, H.; et al. Nonlinear control of transcription through enhancer–promoter interactions. Nature 2022, 604, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Rinzema, N.J.; Sofiadis, K.; Tjalsma, S.J.D.; Verstegen, M.; Oz, Y.; Valdes-Quezada, C.; Felder, A.-K.; Filipovska, T.; van der Elst, S.; de Andrade Dos Ramos, Z.; et al. Building regulatory landscapes reveals that an enhancer can recruit cohesin to create contact domains, engage CTCF sites and activate distant genes. Nat. Struct. Mol. Biol. 2022, 29, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J.-M. DNA loop extrusion by human cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef]
- Vian, L.; Pękowska, A.; Rao, S.S.; Kieffer-Kwon, K.-R.; Jung, S.; Baranello, L.; Huang, S.-C.; El Khattabi, L.; Dose, M.; Pruett, N.; et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell 2018, 173, 1165–1178.e1120. [Google Scholar] [CrossRef]
- Hsieh, T.-H.S.; Cattoglio, C.; Slobodyanyuk, E.; Hansen, A.S.; Rando, O.J.; Tjian, R.; Darzacq, X. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell 2020, 78, 539–553.e538. [Google Scholar] [CrossRef]
- Chen, L.-F.; Long, H.K.; Park, M.; Swigut, T.; Boettiger, A.N.; Wysocka, J. Structural elements promote architectural stripe formation and facilitate ultra-long-range gene regulation at a human disease locus. Mol. Cell 2023, 83, 1446–1461.e6. [Google Scholar] [CrossRef]
- Hsieh, T.-H.S.; Cattoglio, C.; Slobodyanyuk, E.; Hansen, A.S.; Darzacq, X.; Tjian, R. Enhancer–promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat. Genet. 2022, 54, 1919–1932. [Google Scholar] [CrossRef]
- Calderon, L.; Weiss, F.D.; A Beagan, J.; Oliveira, M.S.; Georgieva, R.; Wang, Y.-F.; Carroll, T.S.; Dharmalingam, G.; Gong, W.; Tossell, K.; et al. Cohesin-dependence of neuronal gene expression relates to chromatin loop length. Elife 2022, 11, e76539. [Google Scholar] [CrossRef]
- Aljahani, A.; Hua, P.; Karpinska, M.A.; Quililan, K.; Davies, J.O.J.; Oudelaar, A.M. Analysis of sub-kilobase chromatin topology reveals nano-scale regulatory interactions with variable dependence on cohesin and CTCF. Nat. Commun. 2022, 13, 2139. [Google Scholar] [CrossRef] [PubMed]
- Cuartero, S.; Weiss, F.D.; Dharmalingam, G.; Guo, Y.; Ing-Simmons, E.; Masella, S.; Robles-Rebollo, I.; Xiao, X.; Wang, Y.-F.; Barozzi, I.; et al. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat. Immunol. 2018, 19, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Khrameeva, E.E.; Gavrilov, A.A.; Flyamer, I.M.; Kos, P.; Mikhaleva, E.A.; Penin, A.A.; Logacheva, M.D.; Imakaev, M.V.; Chertovich, A.; et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016, 26, 70–84. [Google Scholar] [CrossRef]
- Rowley, M.J.; Corces, V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018, 19, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Comoglio, F.; Park, H.J.; Schoenfelder, S.; Barozzi, I.; Bode, D.; Fraser, P.; Green, A.R. Thrombopoietin signaling to chromatin elicits rapid and pervasive epigenome remodeling within poised chromatin architectures. Genome Res. 2018, 28, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Ghavi-Helm, Y.; Klein, F.A.; Pakozdi, T.; Ciglar, L.; Noordermeer, D.; Huber, W.; Furlong, E.E.M. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 2014, 512, 96–100. [Google Scholar] [CrossRef]
- Lettice, L.A.; Heaney, S.J.; Purdie, L.A.; Li, L.; De Beer, P.; Oostra, B.A.; Goode, D.; Elgar, G.; Hill, R.E.; De Graaff, E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003, 12, 1725–1735. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Fraser, P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455. [Google Scholar] [CrossRef]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef]
- Razin, S.V.; Ulianov, S.V. Gene functioning and storage within a folded genome. Cell. Mol. Biol. Lett. 2017, 22, 18. [Google Scholar] [CrossRef]
- Valton, A.-L.; Dekker, J. TAD disruption as oncogenic driver. Curr. Opin. Genet. Dev. 2016, 36, 34–40. [Google Scholar] [CrossRef]
- Degl’innocenti, A.; D’errico, A. Regulatory Features for Odorant Receptor Genes in the Mouse Genome. Front. Genet. 2017, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; Kadonaga, J.T. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001, 15, 2515–2519. [Google Scholar] [CrossRef] [PubMed]
- Zabidi, M.A.; Stark, A. Regulatory Enhancer–Core-Promoter Communication via Transcription Factors and Cofactors. Trends Genet. 2016, 32, 801–814. [Google Scholar] [CrossRef]
- Merli, C.; Bergstrom, D.E.; Cygan, J.A.; Blackman, R.K. Promoter specificity mediates the independent regulation of neighbouring genes. Genes Dev. 1996, 10, 1260–1270. [Google Scholar] [CrossRef]
- Martinez-Ara, M.; Comoglio, F.; van Arensbergen, J.; van Steensel, B. Systematic analysis of intrinsic enhancer-promoter compatibility in the mouse genome. Mol. Cell 2022, 82, 2519–2531.e2516. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Ioudinkova, E.S.; Kantidze, O.L.; Iarovaia, O.V. Co-Regulated Genes and Gene Clusters. Genes 2021, 12, 907. [Google Scholar] [CrossRef]
- Tanimoto, K.; Liu, Q.; Bungert, J.; Engel, J.D. Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature 1999, 398, 344–348. [Google Scholar] [CrossRef]
- Stamatoyannopoulos, G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005, 33, 259–271. [Google Scholar] [CrossRef]
- Guo, Y.; Monahan, K.; Wu, H.; Gertz, J.; Varley, K.E.; Li, W.; Myers, R.M.; Maniatis, T.; Wu, Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc. Natl. Acad. Sci. USA 2012, 109, 21081–21086. [Google Scholar] [CrossRef]
- Canzio, D.; Nwakeze, C.L.; Horta, A.; Rajkumar, S.M.; Coffey, E.L.; Duffy, E.E.; Duffié, R.; Monahan, K.; O’keeffe, S.; Simon, M.D.; et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 2019, 177, 639–653.e615. [Google Scholar] [CrossRef]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Brandão, H.B.; Ulianov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.A.; Tachibana-Konwalski, K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Zakharova, V.V.; Galitsyna, A.A.; Kos, P.I.; Polovnikov, K.E.; Flyamer, I.M.; Mikhaleva, E.A.; Khrameeva, E.E.; Germini, D.; Logacheva, M.D.; et al. Order and stochasticity in the folding of individual Drosophila genomes. Nat. Commun. 2021, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Stevens, T.J.; Lando, D.; Basu, S.; Atkinson, L.P.; Cao, Y.; Lee, S.F.; Leeb, M.; Wohlfahrt, K.J.; Boucher, W.; O’shaughnessy-Kirwan, A.; et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 2017, 544, 59–64. [Google Scholar] [CrossRef]
- Tan, L.; Xing, D.; Chang, C.-H.; Li, H.; Xie, X.S. Three-dimensional genome structures of single diploid human cells. Science 2018, 361, 924–928. [Google Scholar] [CrossRef]
- Yu, M.; Abnousi, A.; Zhang, Y.; Li, G.; Lee, L.; Chen, Z.; Fang, R.; Lagler, T.M.; Yang, Y.; Wen, J.; et al. SnapHiC: A computational pipeline to identify chromatin loops from single-cell Hi-C data. Nat. Methods 2021, 18, 1056–1059. [Google Scholar] [CrossRef]
- Zhang, S.; Plummer, D.; Lu, L.; Cui, J.; Xu, W.; Wang, M.; Liu, X.; Prabhakar, N.; Shrinet, J.; Srinivasan, D.; et al. DeepLoop robustly maps chromatin interactions from sparse allele-resolved or single-cell Hi-C data at kilobase resolution. Nat. Genet. 2022, 54, 1013–1025. [Google Scholar] [CrossRef]
- Liu, Q.; Zeng, W.; Zhang, W.; Wang, S.; Chen, H.; Jiang, R.; Zhou, M.; Zhang, S. Deep generative modeling and clustering of single cell Hi-C data. Brief. Bioinform. 2023, 24, bbac494. [Google Scholar] [CrossRef] [PubMed]
- Beliveau, B.J.; Boettiger, A.N.; Avendaño, M.S.; Jungmann, R.; McCole, R.B.; Joyce, E.F.; Kim-Kiselak, C.; Bantignies, F.; Fonseka, C.Y.; Erceg, J.; et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 2015, 6, 7147. [Google Scholar] [CrossRef]
- Bintu, B.; Mateo, L.J.; Su, J.-H.; Sinnott-Armstrong, N.A.; Parker, M.; Kinrot, S.; Yamaya, K.; Boettiger, A.N.; Zhuang, X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 2018, 362, eaau1783. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-H.; Zheng, P.; Kinrot, S.S.; Bintu, B.; Zhuang, X. Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell 2020, 182, 1641–1659.e1626. [Google Scholar] [CrossRef] [PubMed]
- Narendra, V.; Bulajić, M.; Dekker, J.; Mazzoni, E.O.; Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016, 30, 2657–2662. [Google Scholar] [CrossRef] [PubMed]
- Willemin, A.; Lopez-Delisle, L.; Bolt, C.C.; Gadolini, M.-L.; Duboule, D.; Rodriguez-Carballo, E. Induction of a chromatin boundary in vivo upon insertion of a TAD border. PLoS Genet. 2021, 17, e1009691. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Liu, Y.; Qu, H.; Liu, A.; Sun, P.; Wang, X. The effect of CTCF binding sites destruction by CRISPR/Cas9 on transcription of metallothionein gene family in liver hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2019, 510, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Shao, J.; Mitra, J.; Xiong, F.; D’antonio, M.; Wang, R.; Garcia-Bassets, I.; Ma, Q.; Zhu, X.; Lee, J.-H.; et al. Enhancer release and retargeting activates disease-susceptibility genes. Nature 2021, 595, 735–740. [Google Scholar] [CrossRef]
- Kubo, N.; Ishii, H.; Xiong, X.; Bianco, S.; Meitinger, F.; Hu, R.; Hocker, J.D.; Conte, M.; Gorkin, D.; Yu, M.; et al. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021, 28, 152–161. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razin, S.V.; Ulianov, S.V.; Iarovaia, O.V. Enhancer Function in the 3D Genome. Genes 2023, 14, 1277. https://doi.org/10.3390/genes14061277
Razin SV, Ulianov SV, Iarovaia OV. Enhancer Function in the 3D Genome. Genes. 2023; 14(6):1277. https://doi.org/10.3390/genes14061277
Chicago/Turabian StyleRazin, Sergey V., Sergey V. Ulianov, and Olga V. Iarovaia. 2023. "Enhancer Function in the 3D Genome" Genes 14, no. 6: 1277. https://doi.org/10.3390/genes14061277
APA StyleRazin, S. V., Ulianov, S. V., & Iarovaia, O. V. (2023). Enhancer Function in the 3D Genome. Genes, 14(6), 1277. https://doi.org/10.3390/genes14061277





