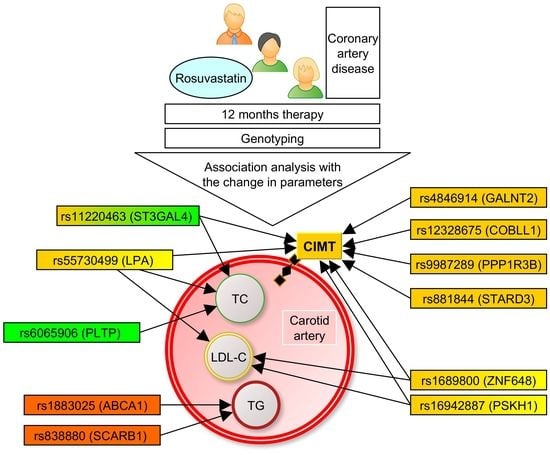
Lipid-Associated GWAS Loci Predict Antiatherogenic Effects of Rosuvastatin in Patients with Coronary Artery Disease
Abstract
1. Introduction
2. Materials and Methods
3. Results
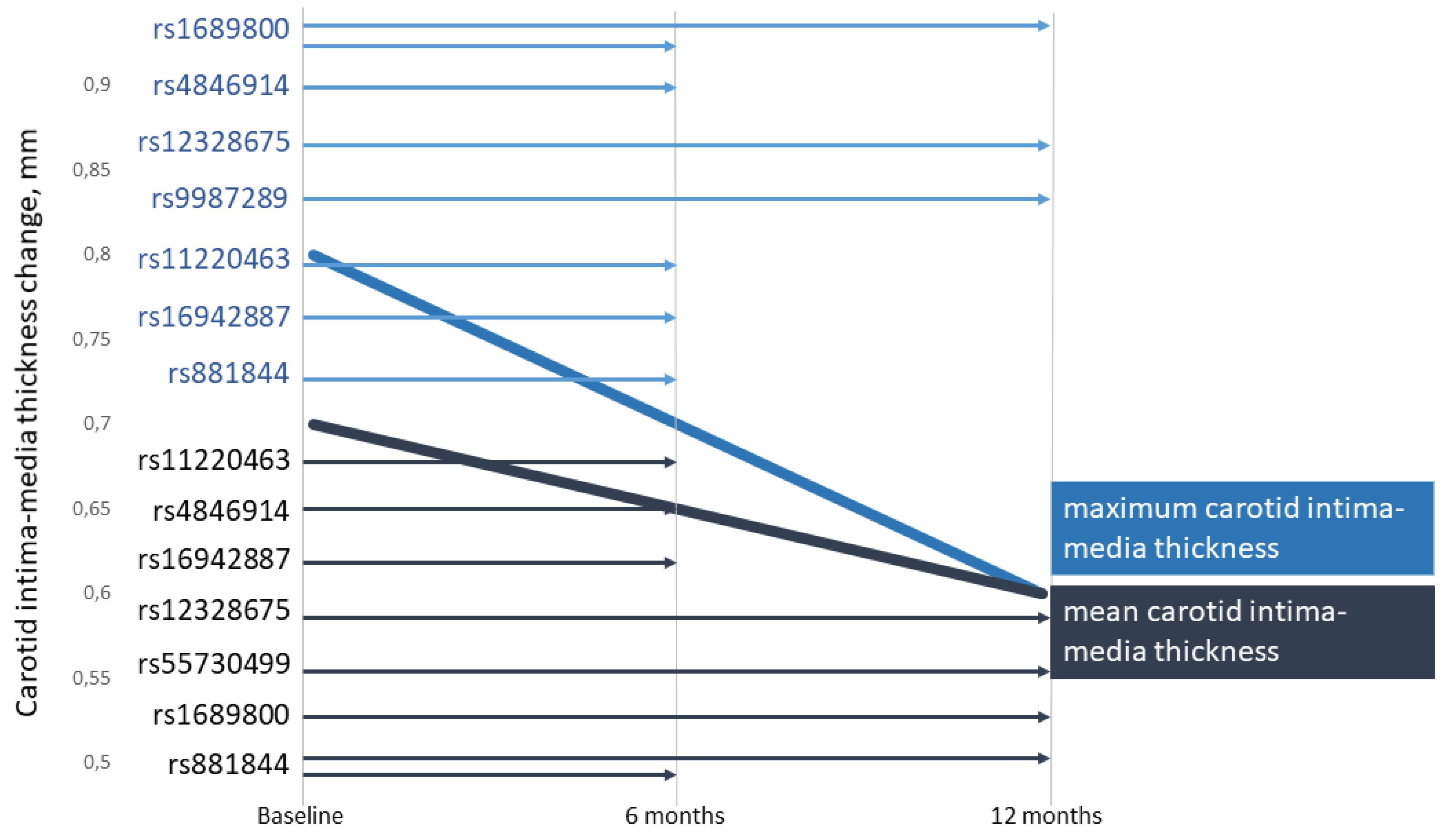
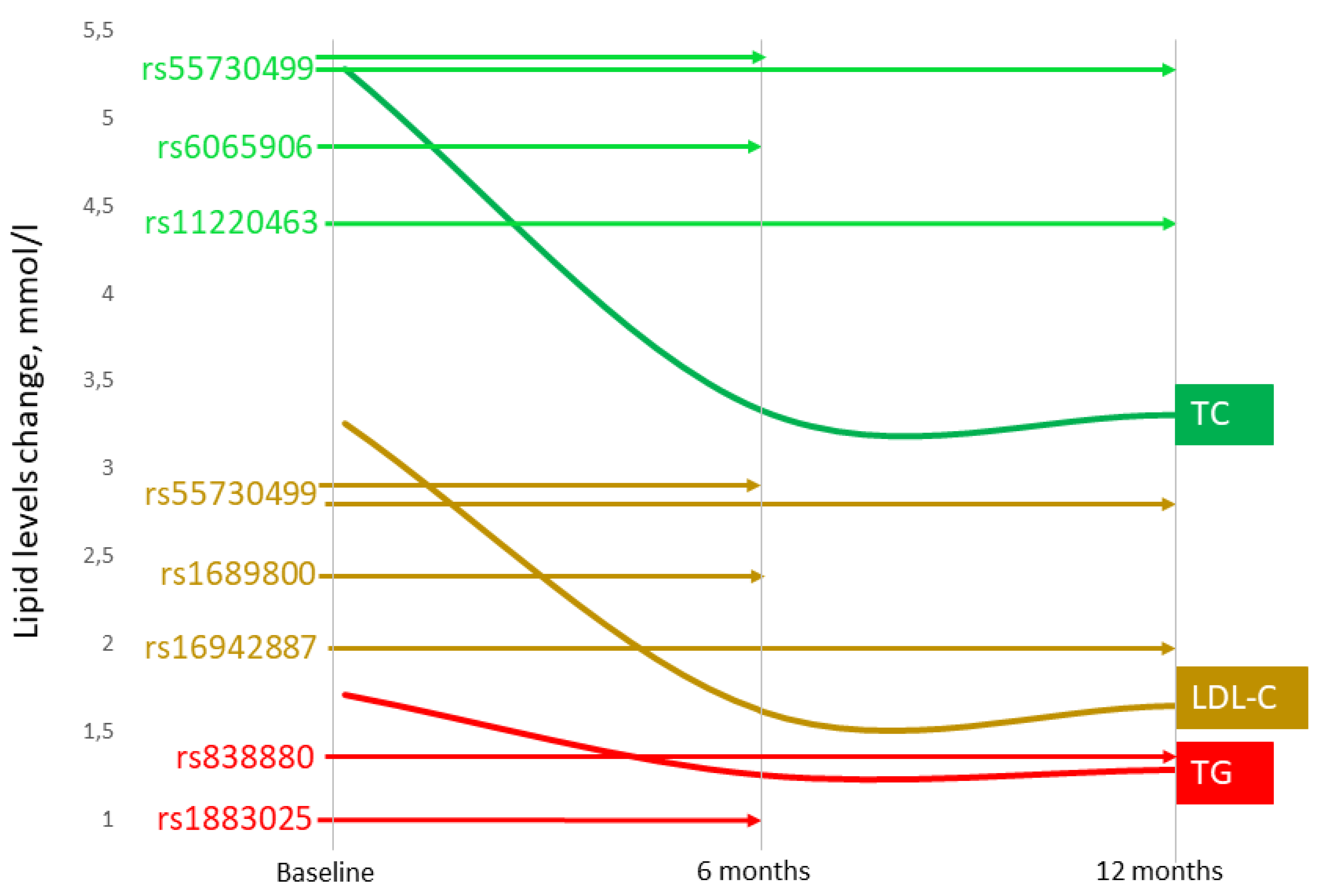
3.1. Associations of the SNPs with Lipid and CIMT Reduction during the 6-Month Therapy by Rosuvastatin
3.2. Associations of SNPs with Lipid and CIMT Reduction during 12-Month Therapy by Rosuvastatin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, R.M.; Schnitzler, G.; Fang, S.; Lee-Kim, V.S.; Barry, A. Multiomic Analysis and CRISPR Perturbation Screens Identify Endothelial Cell Programs and Novel Therapeutic Targets for Coronary Artery Disease. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, H. Invited commentary: 30-year perspective on the seven countries study. Am. J. Epidemiol. 2017, 185, 1143–1147. [Google Scholar] [CrossRef]
- McPherson, R.; Tybjaerg-Hansen, A. Genetics of Coronary Artery Disease. Circ. Res. 2016, 118, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Churilin, M.I.; Kononov, S.I.; Mal, G.S.; Polonikov, A.V.; Lazarenko, V.A. Lipid metabolism genes and predisposition to ischemic heart disease. Med. News North Cauc. 2019, 14, 401–407. [Google Scholar] [CrossRef]
- Mutlu, A.S.; Duffy, J.; Wang, M.C. Lipid metabolism and lipid signals in aging and longevity. Dev. Cell. 2021, 56, 1394–1407. [Google Scholar] [CrossRef]
- Walsh, R.; Jurgens, S.J.; Erdmann, J.; Bezzina, C.R. Genome-wide association studies of cardiovascular disease. Physiol. Rev. 2023, 103, 2039–2055. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [PubMed]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef]
- Kathiresan, S.; Melander, O.; Guiducci, C.; Surti, A.; Burtt, N.P.; Rieder, M.J.; Cooper, G.M.; Roos, C.; Voight, B.F.; Havulinna, A.S.; et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008, 40, 189–197. [Google Scholar] [CrossRef]
- Mack, S.; Coassin, S.; Rueedi, R.; Yousri, N.A.; Seppälä, I.; Gieger, C.; Schönherr, S.; Forer, L.; Erhart, G.; Marques-Vidal, P.; et al. A genome-wide association meta-analysis on lipoprotein (a) concentrations adjusted for apolipoprotein (a) isoforms. J. Lipid Res. 2017, 58, 1834–1844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gan, W.; Tian, C.; Li, H.; Lin, X.; Chen, Y. Association of PPP1R3B polymorphisms with blood lipid and C-reactive protein levels in a Chinese population. J. Diabetes 2013, 5, 275–281. [Google Scholar] [CrossRef]
- Postmus, I.; Trompet, S.; Deshmukh, H.A.; Arsenault, B.J.; Avery, C.L.; Bis, J.C.; Chasman, D.I.; de Keyser, C.E.; Deshmukh, H.A.; Evans, D.S.; et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat. Commun. 2014, 5, 5068. [Google Scholar] [CrossRef] [PubMed]
- Chasman, D.I.; Giulianini, F.; MacFadyen, J.; Barratt, B.J.; Nyberg, F.; Ridker, P.M. Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: The Justification for the Use of Statins in Prevention: An Intervention Trial Evaluating Rosuvastatin (JUPITER) trial. Circ. Cardiovasc. Genet. 2012, 5, 257–264. [Google Scholar] [CrossRef]
- Chasman, D.I.; Giulianini, F.; Demler, O.V.; Udler, M.S. Pleiotropy-Based Decomposition of Genetic Risk Scores: Association and Interaction Analysis for Type 2 Diabetes and CAD. Am. J. Hum. Genet. 2020, 106, 646–658. [Google Scholar] [CrossRef]
- Crouse, J.R., 3rd; Raichlen, J.S.; Riley, W.A.; Evans, G.W.; Palmer, M.K.; O’Leary, D.H.; Grobbee, D.E.; Bots, M.L.; METEOR Study Group. Effect of rosuvastatin on progression of carotid intima-media thickness in low-risk individuals with subclinical atherosclerosis: The METEOR Trial. JAMA 2007, 297, 1344–1353. [Google Scholar] [CrossRef]
- Nezu, T.; Hosomi, N.; Aoki, S.; Matsumoto, M. Carotid Intima-Media Thickness for Atherosclerosis. J. Atheroscler. Thromb. 2016, 23, 18–31. [Google Scholar] [CrossRef]
- Shah, S.; Casas, J.P.; Drenos, F.; Whittaker, J.; Deanfield, J.; Swerdlow, D.I.; Holmes, M.V.; Kivimaki, M.; Langenberg, C.; Wareham, N.; et al. Causal relevance of blood lipid fractions in the development of carotid atherosclerosis: Mendelian randomization analysis. Circ. Cardiovasc. Genet. 2013, 6, 63–72. [Google Scholar] [CrossRef]
- Vargas, J.D.; Manichaikul, A.; Wang, X.Q.; Rich, S.S.; Rotter, J.I.; Post, W.S.; Polak, J.F.; Budoff, M.J.; Bluemke, D.A. Detailed analysis of association between common single nucleotide polymorphisms and subclinical atherosclerosis: The Multi-ethnic Study of Atherosclerosis. Data Brief. 2016, 7, 229–242. [Google Scholar] [CrossRef]
- Guaricci, A.I.; Arcadi, T.; Brunetti, N.D.; Maffei, E.; Montrone, D.; Martini, C.; De Luca, M.; De Rosa, F.; Cocco, D.; Midiri, M.; et al. Carotid intima media thickness and coronary atherosclerosis linkage in symptomatic intermediate risk patients evaluated by coronary computed tomography angiography. Int. J. Cardiol. 2014, 176, 988–993. [Google Scholar] [CrossRef]
- Amato, M.; Montorsi, P.; Ravani, A.; Oldani, E.; Galli, S.; Ravagnani, P.M.; Tremoli, E.; Baldassarre, D. Carotid intima-media thickness by B-mode ultrasound as surrogate of coronary atherosclerosis: Correlation with quantitative coronary angiography and coronary intravascular ultrasound findings. Eur. Heart J. 2007, 28, 2094–2101. [Google Scholar] [CrossRef]
- Granér, M.; Varpula, M.; Kahri, J.; Salonen, R.M.; Nyyssönen, K.; Nieminen, M.S.; Taskinen, M.R.; Syvänne, M. Association of carotid intima-media thickness with angiographic severity and extent of coronary artery disease. Am. J. Cardiol. 2006, 97, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, Y.; Kawano, H.; Koide, Y.; Baba, T.; Toda, G.; Seto, S.; Yano, K. Relationship of carotid intima-media thickness, pulse wave velocity, and ankle brachial index to the severity of coronary artery atherosclerosis. Clin Cardiol. 2004, 27, 629–634. [Google Scholar] [CrossRef]
- Ballantyne, C.M.; Raichlen, J.S.; Nicholls, S.J.; Erbel, R.; Tardif, J.C.; Brener, S.J.; Cain, V.A.; Nissen, S.E.; ASTEROID Investigators. Effect of rosuvastatin therapy on coronary artery stenoses assessed by quantitative coronary angiography: A study to evaluate the effect of rosuvastatin on intravascular ultrasound-derived coronary atheroma burden. Circulation 2008, 117, 2458–2466. [Google Scholar] [CrossRef]
- Puri, R.; Libby, P.; Nissen, S.E.; Wolski, K.; Ballantyne, C.M.; Barter, P.J.; Chapman, M.J.; Erbel, R.; Raichlen, J.S.; Uno, K.; et al. Long-term effects of maximally intensive statin therapy on changes in coronary atheroma composition: Insights from SATURN. Eur. Heart J. Cardiovasc. Imaging 2014, 15, 380–388. [Google Scholar] [CrossRef]
- Rumyantsev, N.A.; Kukes, V.G.; Kazakov, R.E.; Rumyantsev, A.A.; Sychev, D.A. Ispol’zovanie farmakogeneticheskogo testirovaniia dlia predotvrashcheniia nezhelatel’nykh lekarstvennykh reaktsiĭ pri terapii statinami [Use of pharmacogenetic testing to prevent adverse drug reactions during statin therapy]. Ter Arkh. 2017, 89, 82–87. (In Russian) [Google Scholar] [CrossRef]
- Soko, N.D.; Masimirembwa, C.; Dandara, C. Pharmacogenomics of Rosuvastatin: A Glocal (Global + Local) African Perspective and Expert Review on a Statin Drug. OMICS A J. Integr. Biol. 2016, 20, 498–509. [Google Scholar] [CrossRef]
- Alfonsi, J.E.; Hegele, R.A.; Gryn, S.E. Pharmacogenetics of lipid-lowering agents: Precision or indecision medicine? Curr. Atheroscler. Rep. 2016, 18, 24. [Google Scholar] [CrossRef]
- Churilin, M.I.; Kononov, S.I.; Luneva, Y.V.; Kazanov, V.A.; Azarova, I.E.; Klyosova, E.Y.; Bykanova, M.A.; Paschoalini, G.; Kharchenko, A.V.; Zhabin, S.N.; et al. Polymorphisms of Intracellular Cholesterol Transporters Genes: Relationship to Blood Lipid Levels, Carotid Intima-Media Thickness, and the Development of Coronary Heart Disease. Russ. J. Genet. 2020, 56, 234–241. [Google Scholar] [CrossRef]
- Churilin, M.I.; Kononov, S.I.; Luneva, Y.V.; Azarova, Y.E.; Klesova, E.Y.; Kharchenko, A.V.; Zhabin, S.N.; Bushueva, O.Y.; Povetkin, S.V.; Mal, G.S.; et al. Apolipoprotein E gene polymorphisms: A relationship with the risk of coronary artery disease and the effectiveness of lipid-lowering therapy with rosuvastatin. Cardiovasc. Therapy Prev. Russ. Fed. 2020, 19, 17–23. [Google Scholar] [CrossRef]
- Kononov, S.; Mal, G.; Azarova, I.; Klyosova, E.; Bykanova, M.; Churnosov, M.; Polonikov, A. Pharmacogenetic loci for rosuvastatin are associated with intima-media thickness change and coronary artery disease risk. Pharmacogenomics 2022, 23, 15–34. [Google Scholar] [CrossRef]
- Lazarenko, V.; Churilin, M.; Azarova, I.; Klyosova, E.; Bykanova, M.; Ob’edkova, N.; Churnosov, M.; Bushueva, O.; Mal, G.; Povetkin, S.; et al. Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease. Biomedicines 2022, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Pignoli, P.; Tremoli, E.; Poli, A.; Oreste, P.; Paoletti, R. Intimal plus medial thickness of the arterial wall: A direct measurement with ultrasound imaging. Circulation 1986, 74, 1399–1406. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef]
- Shahid, S.U.; Shabana Humphries, S. The SNP rs10911021 is associated with oxidative stress in coronary heart disease patients from Pakistan. Lipids Health Dis. 2018, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Khetarpal, S.A.; Schjoldager, K.T.; Christoffersen, C.; Raghavan, A.; Edmondson, A.C.; Reutter, H.M.; Ahmed, B.; Ouazzani, R.; Peloso, G.M.; Vitali, C.; et al. Loss of Function of GALNT2 Lowers High-Density Lipoproteins in Humans, Nonhuman Primates, and Rodents. Cell Metab. 2016, 24, 234–245. [Google Scholar] [CrossRef]
- Zilmer, M.; Edmondson, A.C.; Khetarpal, S.A.; Alesi, V.; Zaki, M.S.; Rostasy, K.; Madsen, C.G.; Lepri, F.R.; Sinibaldi, L.; Cusmai, R.; et al. Novel congenital disorder of O-linked glycosylation caused by GALNT2 loss of function. Brain 2020, 143, 1114–1126. [Google Scholar] [CrossRef]
- Chen, J.; Guan, L.; Liu, H.; Liu, Q.; Fan, P.; Bai, H. GALNT2 Gene Variant rs4846914 Is Associated with Insulin and Insulin Resistance Depending on BMI in PCOS Patients: A Case-Control Study. Reprod Sci. 2021, 28, 1122–1132. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, H.; Zhang, J.; Han, D.; Zheng, Y.; Guo, X.; He, D.; Guo, J.; Wang, Y. Gene environment interaction of GALNT2 and APOE gene with hypertension in the Chinese Han Population. Biomed. Mater. Eng. 2015, 26 (Suppl. S1), S1977–S2083. [Google Scholar] [CrossRef]
- Guo, Z.; Neilson, L.J.; Zhong, H.; Murray, P.S.; Zanivan, S.; Zaidel-Bar, R. E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci. Signal. 2014, 7, rs7. [Google Scholar] [CrossRef]
- Churilin, M.I. Association of RS12328675 COBLL1 polymorphism with coronary heart disease and intermediate phenotypes of atherosclerosis: Validation study in Central Russia. Res. Results Biomed. 2020, 6, 209–218. [Google Scholar] [CrossRef]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef]
- Mishra, A.; Malik, R.; Hachiya, T.; Jürgenson, T.; Namba, S.; Posner, D.C.; Kamanu, F.K.; Koido, M.; Le Grand, Q.; Shi, M.; et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 2022, 611, 115–123, Erratum in Nature 2022, 612, E7. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef]
- Sethwala, A.M.; Goh, I.; Amerena, J.V. Combating Inflammation in Cardiovascular Disease. Heart Lung Circ. 2021, 30, 197–206. [Google Scholar] [CrossRef]
- Lind, L. Genome-Wide Association Study of the Metabolic Syndrome in UK Biobank. Metab. Syndr. Relat. Disord. 2019, 17, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Spracklen, C.N.; Marenne, G.; Varshney, A.; Corbin, L.J.; Luan, J.; Willems, S.M.; Wu, Y.; Zhang, X.; Horikoshi, M.; et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021, 53, 840–860. [Google Scholar] [CrossRef] [PubMed]
- Meza, C.A.; La Favor, J.D.; Kim, D.H.; Hickner, R.C. Endothelial Dysfunction: Is There a Hyperglycemia-Induced Imbalance of NOX and NOS? Int. J. Mol. Sci. 2019, 20, 3775. [Google Scholar] [CrossRef]
- Clyne, A.M. Endothelial response to glucose: Dysfunction, metabolism, and transport. Biochem. Soc. Trans. 2021, 49, 313–325. [Google Scholar] [CrossRef]
- Krimbou, L.; Denis, M.; Haidar, B.; Carrier, M.; Marcil, M.; Genest, J., Jr. Molecular interactions between apoE and ABCA1: Impact on apoE lipidation. J. Lipid Res. 2004, 45, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.Q.; Sallah, N.; Dunca, D.; Trivedi, B.; Hunt, K.A.; Hodgson, S.; Lambert, S.A.; Arciero, E.; Wright, J.; Griffiths, C.; et al. Transferability of genetic loci and polygenic scores for cardiometabolic traits in British Pakistani and Bangladeshi individuals. Nat. Commun. 2022, 13, 4664. [Google Scholar] [CrossRef]
- Hoffmann, T.J.; Theusch, E.; Haldar, T.; Ranatunga, D.K.; Jorgenson, E.; Medina, M.W.; Kvale, M.N.; Kwok, P.Y.; Schaefer, C.; Krauss, R.M.; et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018, 50, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Byrne, E.M.; Zheng, Z.; Kemper, K.E.; Yengo, L.; Mallett, A.J.; Yang, J.; Visscher, P.M.; Wray, N.R. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat. Commun. 2019, 10, 1891. [Google Scholar] [CrossRef]
- Mondal, N.; Buffone, A., Jr.; Stolfa, G.; Antonopoulos, A.; Lau, J.T.; Haslam, S.M.; Dell, A.; Neelamegham, S. ST3Gal-4 is the primary sialyltransferase regulating the synthesis of E-, P-, and L-selectin ligands on human myeloid leukocytes. Blood 2015, 125, 687–696. [Google Scholar] [CrossRef]
- Ligthart, S.; Vaez, A.; Hsu, Y.H.; Inflammation Working Group of the CHARGE Consortium; PMI-WG-XCP; LifeLines Cohort Study; Stolk, R.; Uitterlinden, A.G.; Hofman, A.; Alizadeh, B.Z.; et al. Bivariate genome-wide association study identifies novel pleiotropic loci for lipids and inflammation. BMC Genom. 2016, 17, 443. [Google Scholar] [CrossRef]
- Ozbeyaz, N.B.; Gokalp, G.; Algul, E.; Kilic, P.; Saricam, O.; Aydinyilmaz, F.; Guliyev, I. Could Systemic Inflammation in Healthy Individuals with Obesity Indicate Subclinical Atherosclerosis? Angiology 2023, 74, 62–69. [Google Scholar] [CrossRef]
- Kivimäki, M.; Lawlor, D.A.; Smith, G.D.; Kumari, M.; Donald, A.; Britton, A.; Casas, J.P.; Shah, T.; Brunner, E.; Timpson, N.J.; et al. Does high C-reactive protein concentration increase atherosclerosis? The Whitehall II Study. PLoS ONE 2008, 3, e3013. [Google Scholar] [CrossRef]
- Moran, C.A.; Sheth, A.N.; Mehta, C.C.; Hanna, D.B.; Gustafson, D.R.; Plankey, M.W.; Mack, W.J.; Tien, P.C.; French, A.L.; Golub, E.T.; et al. The association of C-reactive protein with subclinical cardiovascular disease in HIV-infected and HIV-uninfected women. AIDS 2018, 32, 999–1006. [Google Scholar] [CrossRef]
- Brede, G.; Solheim, J.; Prydz, H. PSKH1, a novel splice factor compartment-associated serine kinase. Nucleic Acids Res. 2002, 30, 5301–5309. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, L.P.; Wendling, C.; Védie, B.; Kobayashi, T.; Chenard, M.P.; Tomasetto, C.; Drin, G.; Alpy, F. STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 2017, 36, 1412–1433. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Y.; Liu, X.; Zheng, N.; Gu, Y.; Song, Y.; Wang, X. Regulatory roles of external cholesterol in human airway epithelial mitochondrial function through STARD3 signalling. Clin. Transl. Med. 2022, 12, e902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhai, X.; Li, J.; Albers, J.J.; Vuletic, S.; Ren, G. Structural basis of the lipid transfer mechanism of phospholipid transfer protein (PLTP). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1082–1094. [Google Scholar] [CrossRef]
- Albers, J.J.; Vuletic, S.; Cheung, M.C. Role of plasma phospholipid transfer protein in lipid and lipoprotein metabolism. Biochim. Biophys. Acta 2012, 1821, 345–357. [Google Scholar] [CrossRef] [PubMed]
| Baseline Parameter | Mean ± Standard Deviation/ Median (Q1; Q3) |
|---|---|
| Age (years) | 61.0 ± 7.25 |
| Body mass index (kg/m2) | 28.77 ± 4.18 |
| Hypertension (%) | 97.5 |
| Past myocardial infarction (%) | 57.6 |
| Systolic blood pressure (mmHg) | 131.1 ± 8.1 |
| Diastolic blood pressure (mmHg) | 74.9 ± 4.4 |
| Total cholesterol (mmol/L) | 5.28 (4.60; 6.06) |
| LDL-C (mmol/L) | 3.27 (2.70; 4.08) |
| HDL-C (mmol/L) | 1.06 (0.97; 1.29) |
| TG (mmol/L) | 1.71 (1.22; 2.37) |
| CIMT, maximum (mm) | 0.80 (0.60; 1.00) |
| CIMT, mean (mm) | 0.70 (0.55; 0.85) |
| Chr | Gene (SNP ID) | Effect Allele | EAF | N | Total Cholesterol | LDL-C | CIMT, Maximum | CIMT, Mean | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta * | Pperm # | Beta * | Pperm # | Beta * | Pperm # | Beta * | Pperm # | |||||
| 1 | ZNF648 (rs1689800) | G | 0.392 | 115 | 0.020 | 0.1786 | 0.046 | 0.0493 A | −0.084 | 0.0234 R | −0.021 | 0.2308 |
| 1 | GALNT2 (rs4846914) | G | 0.388 | 115 | 0.004 | 0.7778 | −0.005 | 0.8571 | −0.045 | 0.0133 A | −0.038 | 0.0344 A |
| 2 | COBLL1 (rs12328675) | C | 0.170 | 111 | −0.003 | 0.8571 | −0.003 | 1.0000 | 0.035 | 0.2647 | 0.041 | 0.1000 |
| 6 | LPA (rs55730499) | T | 0.056 | 115 | 0.323 | 0.0022 R | 0.504 | 0.0224 R | −0.010 | 0.8571 | 0.028 | 0.8571 |
| 7 | NPC1L1 (rs217406) | G | 0.203 | 115 | 0.017 | 0.2308 | 0.033 | 0.3556 | −0.001 | 1.0000 | −0.003 | 1.0000 |
| 8 | PPP1R3B (rs9987289) | A | 0.086 | 115 | 0.019 | 0.3404 | −0.013 | 0.8571 | 0.041 | 0.3404 | 0.051 | 0.1550 |
| 9 | ABCA1 (rs1883025) | T | 0.263 | 115 | 0.001 | 1.0000 | −0.004 | 0.8571 | −0.024 | 0.2982 | −0.025 | 0.2535 |
| 11 | F2 (rs3136441) | C | 0.180 | 102 | 0.011 | 0.6429 | 0.025 | 0.7778 | 0.021 | 0.6923 | 0.023 | 0.2982 |
| 11 | ST3GAL4 (rs11220463) | T | 0.190 | 115 | −0.031 | 0.1280 | −0.068 | 0.0803 | 0.066 | 0.0159 D | 0.061 | 0.0243 A |
| 12 | SCARB1 (rs838880) | C | 0.336 | 115 | 0.002 | 1.0000 | 0.005 | 1.0000 | −0.014 | 0.7273 | −0.015 | 0.6923 |
| 16 | CETP (rs3764261) | A | 0.147 | 115 | 0.007 | 0.6250 | 0.027 | 0.8571 | −0.045 | 0.2466 | −0.042 | 0.1900 |
| 16 | PSKH1 (rs16942887) | A | 0.116 | 115 | 0.009 | 0.6429 | 0.025 | 0.4643 | 0.066 | 0.0421 D | 0.068 | 0.0483 D |
| 17 | STARD3 (rs881844) | C | 0.310 | 115 | 0.003 | 0.8571 | −0.038 | 0.1667 | 0.048 | 0.0086 A | 0.057 | 0.0033 A |
| 19 | LILRA3 (rs386000) | C | 0.203 | 115 | 0.006 | 0.5455 | −0.001 | 1.0000 | 0.015 | 0.7778 | 0.003 | 0.8571 |
| 20 | PLTP (rs6065906) | C | 0.160 | 115 | 0.140 | 0.0135 R | −0.008 | 1.0000 | −0.022 | 0.6923 | −0.022 | 0.5455 |
| Chr | Gene (SNP ID) | Effect Allele | EAF | N | 6-Month Period | 12-Month Period | ||
|---|---|---|---|---|---|---|---|---|
| Beta * | Pperm # | Beta * | Pperm # | |||||
| 1 | ZNF648 (rs1689800) | G | 0.392 | 114 | −0.6495 | 0.1148 | −0.1946 | 0.4643 |
| 1 | GALNT2 (rs4846914) | G | 0.388 | 114 | 0.4816 | 0.1919 | 0.05046 | 0.7778 |
| 2 | COBLL1 (rs12328675) | C | 0.170 | 110 | −0.2948 | 0.6250 | −0.1431 | 0.7778 |
| 6 | LPA (rs55730499) | T | 0.056 | 114 | −0.5923 | 0.4242 | 0.1825 | 0.6923 |
| 7 | NPC1L1 (rs217406) | G | 0.203 | 114 | 0.4106 | 0.4118 | −0.1208 | 0.6429 |
| 8 | PPP1R3B (rs9987289) | A | 0.086 | 114 | −0.9197 | 0.1887 | −0.7977 | 0.0756 |
| 9 | ABCA1 (rs1883025) | T | 0.263 | 114 | −0.177 | 0.5789 | −0.7246 | 0.0160 D |
| 11 | F2 (rs3136441) | C | 0.180 | 101 | −0.185 | 0.5789 | 0.02512 | 1.0000 |
| 11 | ST3GAL4 (rs11220463) | T | 0.190 | 114 | −0.774 | 0.1587 | 3.624 | 0.0503 R |
| 12 | SCARB1 (rs838880) | C | 0.336 | 114 | 1.736 | 0.0478 R | 0.1063 | 0.6429 |
| 16 | CETP (rs3764261) | A | 0.147 | 114 | 0.695 | 0.2931 | 0.02338 | 1.0000 |
| 16 | PSKH1 (rs16942887) | A | 0.116 | 114 | −0.6306 | 0.1439 | −0.2626 | 0.3947 |
| 17 | STARD3 (rs881844) | C | 0.310 | 114 | 0.4075 | 0.4815 | −0.2337 | 0.2647 |
| 19 | LILRA3 (rs386000) | C | 0.203 | 114 | −0.1847 | 0.8571 | −0.1448 | 0.6923 |
| 20 | PLTP (rs6065906) | C | 0.160 | 114 | −0.4654 | 0.5789 | −0.0718 | 0.8571 |
| Chr | Gene (SNP ID) | Effect Allele | EAF | N | Total Cholesterol | LDL-C | CIMT, Maximum | CIMT, Mean | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta * | Pperm # | Beta * | Pperm # | Beta * | Pperm # | Beta * | Pperm # | |||||
| 1 | ZNF648 (rs1689800) | G | 0.392 | 113 | 0.010 | 0.3091 | 0.024 | 0.2043 | −0.093 | 0.0105 R | −0.036 | 0.0282 A |
| 1 | GALNT2 (rs4846914) | G | 0.388 | 113 | −0.004 | 0.7273 | −0.016 | 0.4118 | 0.002 | 0.8571 | −0.002 | 0.8571 |
| 2 | COBLL1 (rs12328675) | C | 0.170 | 109 | −0.011 | 0.4643 | −0.018 | 0.4643 | 0.059 | 0.0213 A | 0.075 | 0.0056 D |
| 6 | LPA (rs55730499) | T | 0.056 | 113 | 0.364 | 0.0001 R | 0.367 | 0.0415 R | 0.018 | 0.8571 | 0.414 | 0.0146 R |
| 7 | NPC1L1 (rs217406) | G | 0.203 | 113 | −0.002 | 1.0000 | 0.003 | 0.2043 | 0.019 | 0.3636 | 0.018 | 0.4516 |
| 8 | PPP1R3B (rs9987289) | A | 0.086 | 113 | 0.004 | 0.8571 | −0.068 | 0.4118 | 0.072 | 0.0359 D | 0.079 | 0.0109 D |
| 9 | ABCA1 (rs1883025) | T | 0.263 | 113 | 0.010 | 0.3148 | 0.001 | 0.4643 | 0.011 | 0.8571 | 0.019 | 0.2687 |
| 11 | F2 (rs3136441) | C | 0.180 | 100 | 0.013 | 0.3478 | 0.018 | 0.2043 | −0.011 | 0.6429 | −0.005 | 1.0000 |
| 11 | ST3GAL4 (rs11220463) | T | 0.190 | 113 | −0.181 | 0.0273 R | −0.027 | 0.4118 | 0.001 | 1.0000 | 0.032 | 0.1709 |
| 12 | SCARB1 (rs838880) | C | 0.336 | 113 | −0.001 | 1.0000 | −0.005 | 0.4643 | −0.003 | 1.0000 | −0.004 | 1.0000 |
| 16 | CETP (rs3764261) | A | 0.147 | 113 | 0.017 | 0.3478 | 0.021 | 0.2043 | −0.053 | 0.0833 | −0.043 | 0.0880 |
| 16 | PSKH1 (rs16942887) | A | 0.116 | 113 | 0.006 | 0.5789 | 0.086 | 0.0175 D | −0.011 | 0.5217 | −0.004 | 0.7778 |
| 17 | STARD3 (rs881844) | C | 0.310 | 113 | 0.009 | 0.4375 | 0.010 | 0.8571 | 0.028 | 0.1852 | 0.040 | 0.0223 A |
| 19 | LILRA3 (rs386000) | C | 0.203 | 113 | 0.019 | 0.2400 | 0.015 | 0.5200 | 0.026 | 0.2982 | 0.010 | 0.7778 |
| 20 | PLTP (rs6065906) | C | 0.160 | 113 | 0.013 | 0.4815 | 0.031 | 0.2571 | −0.029 | 0.3478 | −0.006 | 1.0000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kononov, S.; Azarova, I.; Klyosova, E.; Bykanova, M.; Churnosov, M.; Solodilova, M.; Polonikov, A. Lipid-Associated GWAS Loci Predict Antiatherogenic Effects of Rosuvastatin in Patients with Coronary Artery Disease. Genes 2023, 14, 1259. https://doi.org/10.3390/genes14061259
Kononov S, Azarova I, Klyosova E, Bykanova M, Churnosov M, Solodilova M, Polonikov A. Lipid-Associated GWAS Loci Predict Antiatherogenic Effects of Rosuvastatin in Patients with Coronary Artery Disease. Genes. 2023; 14(6):1259. https://doi.org/10.3390/genes14061259
Chicago/Turabian StyleKononov, Stanislav, Iuliia Azarova, Elena Klyosova, Marina Bykanova, Mikhail Churnosov, Maria Solodilova, and Alexey Polonikov. 2023. "Lipid-Associated GWAS Loci Predict Antiatherogenic Effects of Rosuvastatin in Patients with Coronary Artery Disease" Genes 14, no. 6: 1259. https://doi.org/10.3390/genes14061259
APA StyleKononov, S., Azarova, I., Klyosova, E., Bykanova, M., Churnosov, M., Solodilova, M., & Polonikov, A. (2023). Lipid-Associated GWAS Loci Predict Antiatherogenic Effects of Rosuvastatin in Patients with Coronary Artery Disease. Genes, 14(6), 1259. https://doi.org/10.3390/genes14061259