Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Genotyping Quality Control
2.2. Genome-Wide Association Studies
2.3. Selection Signature Detection Using the iHS Method
2.4. ROH Detection
3. Results
3.1. Genome-Wide Association Studies and Annotation of Candidate Genes
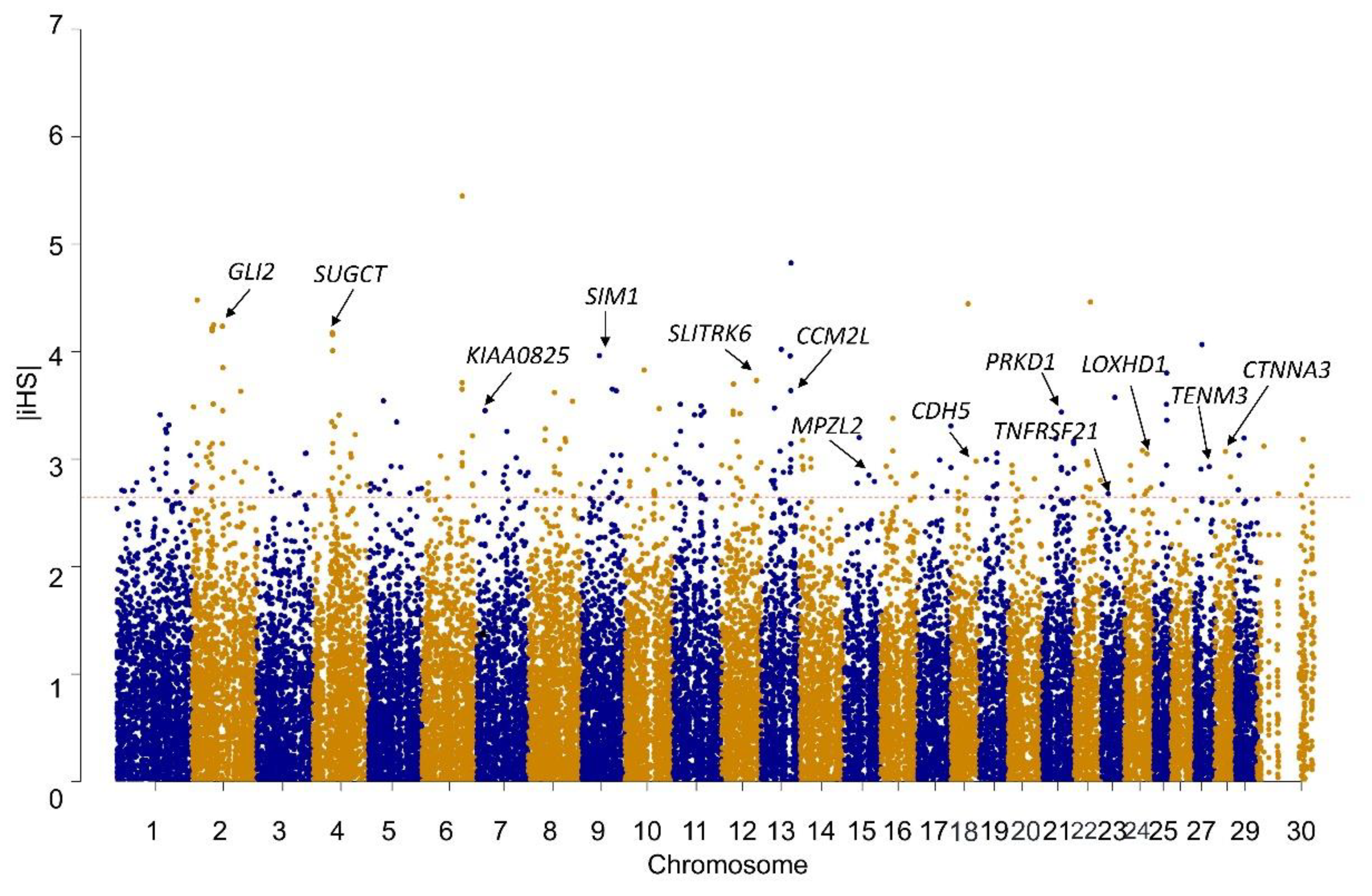
3.2. Selection Signatures Detected by iHS
3.3. ROH Results
3.3.1. Genomic ROH Distribution
3.3.2. Genomic Patterns of ROHs
3.3.3. The Consensus ROH Regions and the Genes Overlaping with Those Detected on the Basis of iHS
4. Discussion
4.1. Litter Size
4.2. Coat Colour
4.3. Black Middorsal Stripe
4.4. Skin Colour
4.5. Selection Signatures in YZD Goats
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brito, L.; Kijas, J.; Ventura, R.; Sargolzaei, M.; Porto-Neto, L.; Cánovas, A.; Feng, Z.; Jafarikia, M.; Schenkel, F. Genetic diversity and signatures of selection in various goat breeds revealed by genome-wide SNP markers. BMC Genom. 2017, 18, 229. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.N.; Zhai, H.L.; Cheng, M.; Ma, J.Y.; Cheng, S.F.; Ge, W.; Zhang, G.L.; Wang, J.J.; Zhang, R.Q.; Wang, X.; et al. Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (Capra hircus). Sci. Rep. 2016, 6, 38096. [Google Scholar] [CrossRef] [PubMed]
- Nazari-Ghadikolaei, A.; Mehrabani-Yeganeh, H.; Miarei-Aashtiani, S.; Staiger, E.; Rashidi, A.; Huson, H. Genome-Wide Association Studies Identify Candidate Genes for Coat Color and Mohair Traits in the Iranian Markhoz Goat. Front. Genet. 2018, 9, 105. [Google Scholar] [CrossRef]
- Henkel, J.; Dubacher, A.; Bangerter, E.; Herren, U.; Ammann, P.; Drögemüller, C.; Flury, C.; Leeb, T. Introgression of ASIP and TYRP1 Alleles Explains Coat Color Variation in Valais Goats. J. Hered. 2021, 112, 452–457. [Google Scholar] [CrossRef]
- Ren, H.; Wang, G.; Chen, L.; Jiang, J.; Liu, L.; Li, N.; Zhao, J.; Sun, X.; Zhou, P. Genome-wide analysis of long non-coding RNAs at early stage of skin pigmentation in goats (Capra hircus). BMC Genom. 2016, 17, 67. [Google Scholar] [CrossRef]
- Zhou, P.; Jiang, J.; Huang, Y.; Zhang, L.; Huang, J. Study on Carcass Quality and Meat Traits of Youzhou Black Goats, Local White Goats and Upgrading Offspring of Boer Goat Population. J. Southwest Univ. Nat. Sci. Ed. 2017, 39, 7. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S.; et al. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef]
- Verardo, L.L.; FF, E.S.; Machado, M.A.; do Carmo Panetto, J.C.; de Lima Reis Faza, D.R.; Otto, P.I.; de Almeida Regitano, L.C.; da Silva, L.O.C.; do Egito, A.A.; do Socorro Maues Albuquerque, M.; et al. Genome-Wide Analyses Reveal the Genetic Architecture and Candidate Genes of Indicine, Taurine, Synthetic Crossbreds, and Locally Adapted Cattle in Brazil. Front. Genet. 2021, 12, 702822. [Google Scholar] [CrossRef]
- Signer-Hasler, H.; Henkel, J.; Bangerter, E.; Bulut, Z.; Drögemüller, C.; Leeb, T.; Flury, C. Runs of homozygosity in Swiss goats reveal genetic changes associated with domestication and modern selection. Genet. Sel. Evol. GSE 2022, 54, 6. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.; Schmidt, M.; Savenkova, M.; Sadler-Riggleman, I.; Nilsson, E. Regulation of granulosa and theca cell transcriptomes during ovarian antral follicle development. Mol. Reprod. Dev. 2008, 75, 1457–1472. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, X.; Han, Z.; Wang, D.; Ma, Y.; Liang, C. Loss of CENPF leads to developmental failure in mouse embryos. Cell Cycle Georget. Tex. 2019, 18, 2784–2799. [Google Scholar] [CrossRef] [PubMed]
- Toralova, T.; Susor, A.; Nemcova, L.; Kepkova, K.; Kanka, J. Silencing CENPF in bovine preimplantation embryo induces arrest at 8-cell stage. Reproduction 2009, 138, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; He, X.; Jiang, Y.; Liu, Y.; Ouyang, Y.; Shen, Y.; Hong, Q.; Chu, M. Genome-Wide Analyses Reveal Genetic Convergence of Prolificacy between Goats and Sheep. Genes 2021, 12, 480. [Google Scholar] [CrossRef]
- Adamczak, R.; Ukleja-Sokolowska, N.; Lis, K.; Bartuzi, Z.; Dubiel, M. Progesterone-induced blocking factor 1 and cytokine profile of follicular fluid of infertile women qualified to in vitro fertilization: The influence on fetus development and pregnancy outcome. Int. J. Immunopathol. Pharm. 2022, 36, 3946320221111134. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; He, X.Y.; Wang, F.Y.; Pan, L.X.; Wang, X.Y.; Gan, S.Q.; Di, R.; Chu, M.X. Identification of genes associated with litter size combining genomic approaches in Luzhong mutton sheep. Anim. Genet. 2021, 52, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Hu, W.; Chen, S.; Di, R.; Liu, Q.; Wang, X.; He, X.; Gan, S.; Zhang, X.; Zhang, J.; et al. The genetic mechanism of high prolificacy in small tail han sheep by comparative proteomics of ovaries in the follicular and luteal stages. J. Proteom. 2019, 204, 103394. [Google Scholar] [CrossRef] [PubMed]
- Sell-Kubiak, E.; Dobrzanski, J.; Derks, M.F.L.; Lopes, M.S.; Szwaczkowski, T. Meta-Analysis of SNPs Determining Litter Traits in Pigs. Genes 2022, 13, 1730. [Google Scholar] [CrossRef]
- Hui, Y.; Zhang, Y.; Wang, K.; Pan, C.; Chen, H.; Qu, L.; Song, X.; Lan, X. Goat DNMT3B: An indel mutation detection, association analysis with litter size and mRNA expression in gonads. Theriogenology 2020, 147, 108–115. [Google Scholar] [CrossRef]
- Alfadhel, M.; Albahkali, S.; Almuaysib, A.; Alrfaei, B.M. The SORCS3 gene is mutated in brothers with infantile spasms and intellectual disability. Discov. Med. 2018, 26, 147–153. [Google Scholar]
- Subkhangulova, A.; Malik, A.R.; Hermey, G.; Popp, O.; Dittmar, G.; Rathjen, T.; Poy, M.N.; Stumpf, A.; Beed, P.S.; Schmitz, D.; et al. SORCS1 and SORCS3 control energy balance and orexigenic peptide production. EMBO Rep. 2018, 19, e44810. [Google Scholar] [CrossRef]
- Marchesi, J.A.P.; Ono, R.K.; Cantao, M.E.; Ibelli, A.M.G.; Peixoto, J.O.; Moreira, G.C.M.; Godoy, T.F.; Coutinho, L.L.; Munari, D.P.; Ledur, M.C. Exploring the genetic architecture of feed efficiency traits in chickens. Sci. Rep. 2021, 11, 4622. [Google Scholar] [CrossRef]
- Ding, R.; Zhuang, Z.; Qiu, Y.; Ruan, D.; Wu, J.; Ye, J.; Cao, L.; Zhou, S.; Zheng, E.; Huang, W.; et al. Identify known and novel candidate genes associated with backfat thickness in Duroc pigs by large-scale genome-wide association analysis. J. Anim. Sci. 2022, 100, skac012. [Google Scholar] [CrossRef]
- Shen, J.; Chen, Q.; Zhang, F.; Hanif, Q.; Huang, B.; Chen, N.; Qu, K.; Zhan, J.; Chen, H.; Jiang, Y.; et al. Genome-wide association study identifies quantitative trait loci affecting cattle temperament. Zool. Res. 2022, 43, 14–25. [Google Scholar] [CrossRef]
- Guo, J.; Sun, X.; Mao, A.; Liu, H.; Zhan, S.; Li, L.; Zhong, T.; Wang, L.; Cao, J.; Liu, G.; et al. A 13.42-kb tandem duplication at the ASIP locus is strongly associated with the depigmentation phenotype of non-classic Swiss markings in goats. BMC Genom. 2022, 23, 437. [Google Scholar] [CrossRef]
- Burren, A.; Neuditschko, M.; Signer-Hasler, H.; Frischknecht, M.; Reber, I.; Menzi, F.; Drögemüller, C.; Flury, C. Genetic diversity analyses reveal first insights into breed-specific selection signatures within Swiss goat breeds. Anim. Genet. 2016, 47, 727–739. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhou, G.; Guo, J.; Yan, H.; Niu, Y.; Li, Y.; Yuan, C.; Geng, R.; Lan, X.; et al. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Sci. Rep. 2016, 6, 38932. [Google Scholar] [CrossRef]
- Gao, J.; Lyu, Y.; Zhang, D.; Reddi, K.; Sun, F.; Yi, J.; Liu, C.; Li, H.; Yao, H.; Dai, J.; et al. Genomic Characteristics and Selection Signatures in Indigenous Chongming White Goat (Capra hircus). Front. Genet. 2020, 11, 901. [Google Scholar] [CrossRef]
- Brito, S.; Baek, J.M.; Cha, B.; Heo, H.; Lee, S.H.; Lei, L.; Jung, S.Y.; Lee, S.M.; Lee, S.H.; Kwak, B.M.; et al. Nicotinamide mononucleotide reduces melanin production in aged melanocytes by inhibiting cAMP/Wnt signaling. J. Derm. Sci. 2022, 106, 159–169. [Google Scholar] [CrossRef]
- Wang, X.; Ma, Q. Genome-wide association studies for coat color in Tan sheep. Acta Agric. Zhejiangensis 2020, 32, 7. [Google Scholar] [CrossRef]
- Domenzain-Reyna, C.; Hernandez, D.; Miquel-Serra, L.; Docampo, M.J.; Badenas, C.; Fabra, A.; Bassols, A. Structure and regulation of the versican promoter: The versican promoter is regulated by AP-1 and TCF transcription factors in invasive human melanoma cells. J. Biol. Chem. 2009, 284, 12306–12317. [Google Scholar] [CrossRef]
- Olsson, M.; Durbeej, M.; Ekblom, P.; Hjalt, T. Nulp1, a novel basic helix-loop-helix protein expressed broadly during early embryonic organogenesis and prominently in developing dorsal root ganglia. Cell Tissue Res. 2002, 308, 361–370. [Google Scholar] [CrossRef]
- Fitch, K.R.; McGowan, K.A.; van Raamsdonk, C.D.; Fuchs, H.; Lee, D.; Puech, A.; Herault, Y.; Threadgill, D.W.; Hrabe de Angelis, M.; Barsh, G.S. Genetics of dark skin in mice. Genes Dev. 2003, 17, 214–228. [Google Scholar] [CrossRef]
- Martin, A.R.; Lin, M.; Granka, J.M.; Myrick, J.W.; Liu, X.; Sockell, A.; Atkinson, E.G.; Werely, C.J.; Moller, M.; Sandhu, M.S.; et al. An Unexpectedly Complex Architecture for Skin Pigmentation in Africans. Cell 2017, 171, 1340–1353.e14. [Google Scholar] [CrossRef]
- Shi, X.; Wu, J.; Lang, X.; Wang, C.; Bai, Y.; Riley, D.; Liu, L.; Ma, X. Comparative transcriptome and histological analyses provide insights into the skin pigmentation in Minxian black fur sheep (Ovis aries). PeerJ 2021, 9, e11122. [Google Scholar] [CrossRef]
- Jara, E.; Penagaricano, F.; Armstrong, E.; Ciappesoni, G.; Iriarte, A.; Navajas, E.A. Revealing the genetic basis of eyelid pigmentation in Hereford cattle. J. Anim. Sci. 2022, 100, skac110. [Google Scholar] [CrossRef]
- Wang, F.; Luo, Q.; Chen, Y.; Liu, Y.; Xu, K.; Adhikari, K.; Cai, X.; Liu, J.; Li, Y.; Liu, X.; et al. A Genome-Wide Scan on Individual Typology Angle Found Variants at SLC24A2 Associated with Skin Color Variation in Chinese Populations. J. Investig. Derm. 2022, 142, 1223–1227.e14. [Google Scholar] [CrossRef]
- Hou, H.; Wang, X.; Zhang, C.; Tu, Y.; Lv, W.; Cai, X.; Xu, Z.; Yao, J.; Yang, C. Genomic analysis of GBS data reveals genes associated with facial pigmentation in Xinyang blue-shelled layers. Arch. Anim. Breed. 2020, 63, 483–491. [Google Scholar] [CrossRef]
- Li, D.; Sun, G.; Zhang, M.; Cao, Y.; Zhang, C.; Fu, Y.; Li, F.; Li, G.; Jiang, R.; Han, R.; et al. Breeding history and candidate genes responsible for black skin of Xichuan black-bone chicken. BMC Genom. 2020, 21, 511. [Google Scholar] [CrossRef]
- Bovo, S.; Schiavo, G.; Kazemi, H.; Moscatelli, G.; Ribani, A.; Ballan, M.; Bonacini, M.; Prandi, M.; Dall’Olio, S.; Fontanesi, L. Exploiting within-breed variability in the autochthonous Reggiana breed identified several candidate genes affecting pigmentation-related traits, stature and udder defects in cattle. Anim. Genet. 2021, 52, 579–597. [Google Scholar] [CrossRef]
- Garcia-Gamez, E.; Reverter, A.; Whan, V.; McWilliam, S.M.; Arranz, J.J.; International Sheep Genomics, C.; Kijas, J. Using regulatory and epistatic networks to extend the findings of a genome scan: Identifying the gene drivers of pigmentation in merino sheep. PLoS ONE 2011, 6, e21158. [Google Scholar] [CrossRef]
- Salime, S.; Riahi, Z.; Elrharchi, S.; Elkhattabi, L.; Charoute, H.; Nahili, H.; Rouba, H.; Kabine, M.; Bonnet, C.; Petit, C.; et al. A novel mutation in SLITRK6 causes deafness and myopia in a Moroccan family. Gene 2018, 659, 89–92. [Google Scholar] [CrossRef]
- Pan, H.; Wu, S.; Wang, J.; Zhu, T.; Li, T.; Wan, B.; Liu, B.; Luo, Y.; Ma, X.; Sui, R.; et al. TNFRSF21 mutations cause high myopia. J. Med. Genet 2019, 56, 671–677. [Google Scholar] [CrossRef]
- Swierkowska, J.; Vishweswaraiah, S.; Mrugacz, M.; Radhakrishna, U.; Gajecka, M. Differential methylation of microRNA encoding genes may contribute to high myopia. Front. Genet. 2022, 13, 1089784. [Google Scholar] [CrossRef]
- Fadaie, Z.; Whelan, L.; Ben-Yosef, T.; Dockery, A.; Corradi, Z.; Gilissen, C.; Haer-Wigman, L.; Corominas, J.; Astuti, G.D.N.; de Rooij, L.; et al. Whole genome sequencing and in vitro splice assays reveal genetic causes for inherited retinal diseases. NPJ Genom. Med. 2021, 6, 97. [Google Scholar] [CrossRef]
- Doll, J.; Vona, B.; Schnapp, L.; Ruschendorf, F.; Khan, I.; Khan, S.; Muhammad, N.; Alam Khan, S.; Nawaz, H.; Khan, A.; et al. Genetic Spectrum of Syndromic and Non-Syndromic Hearing Loss in Pakistani Families. Genes 2020, 11, 1329. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, L.; Yang, L.; Wang, L.; Lu, Y.; Dong, X.; Xiao, T.; Xu, Z.; Wu, B.; Zhou, W. Association Between Expanded Genomic Sequencing Combined With Hearing Screening and Detection of Hearing Loss Among Newborns in a Neonatal Intensive Care Unit. JAMA Netw. Open. 2022, 5, e2220986. [Google Scholar] [CrossRef]
- Bademci, G.; Abad, C.; Incesulu, A.; Rad, A.; Alper, O.; Kolb, S.M.; Cengiz, F.B.; Diaz-Horta, O.; Silan, F.; Mihci, E.; et al. MPZL2 is a novel gene associated with autosomal recessive nonsyndromic moderate hearing loss. Hum. Genet. 2018, 137, 479–486. [Google Scholar] [CrossRef]
- Wang, W.Q.; Gao, X.; Huang, S.S.; Kang, D.Y.; Xu, J.C.; Yang, K.; Han, M.Y.; Zhang, X.; Yang, S.Y.; Yuan, Y.Y.; et al. Genetic Analysis of the LOXHD1 Gene in Chinese Patients with Non-Syndromic Hearing Loss. Front. Genet. 2022, 13, 825082. [Google Scholar] [CrossRef]
- Zheng, S.; Xu, P.; Wu, Z.; Zhang, H.; Li, D.; Liu, S.; Liu, B.; Ren, J.; Chen, H.; Huang, M. Genetic structure and domestication footprints of the tusk, coat color, and ear morphology in East Chinese pigs. J. Genet. Genom. 2022, 49, 1053–1063. [Google Scholar] [CrossRef]
- Heydari, R.; Seresht-Ahmadi, M.; Mirshahvaladi, S.; Sabbaghian, M.; Mohseni-Meybodi, A. KIF3B gene silent variant leading to sperm morphology and motility defects and male infertilitydagger. Biol. Reprod. 2022, 106, 766–774. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Gao, X.; Du, C.; Hou, C.; Tang, D.; Lou, B.; Shen, W.; Zhu, J. The potential function of KIF17 in large yellow croaker (Larimichthys crocea) spermatid remodeling: Molecular characterization and expression pattern during spermiogenesis. Fish. Physiol. Biochem. 2022, 48, 603–616. [Google Scholar] [CrossRef]
- Gaitskell-Phillips, G.; Martin-Cano, F.E.; Ortiz-Rodriguez, J.M.; Silva-Rodriguez, A.; da Silva-Alvarez, E.; Gil, M.C.; Ortega-Ferrusola, C.; Pena, F.J. The seminal plasma proteins Peptidyl arginine deaminase 2, rRNA adenine N (6)-methyltransferase and KIAA0825 are linked to better motility post thaw in stallions. Theriogenology 2022, 177, 94–102. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, X.; Yan, Y.; Liu, G.; Liu, C. Expression of cell proliferation regulatory factors bricd5, tnfrsf21, cdk1 correlates with expression of clock gene cry1 in testes of Hu rams during puberty. Mol. Biol. Rep. 2021, 48, 7379–7385. [Google Scholar] [CrossRef]
- Liu, M.; Alharbi, M.; Graves, D.; Yang, S. IFT80 Is Required for Fracture Healing Through Controlling the Regulation of TGF-beta Signaling in Chondrocyte Differentiation and Function. J. Bone Min. Res. 2020, 35, 571–582. [Google Scholar] [CrossRef]
- Wu, M.; Zhao, H.; Tang, X.; Li, Q.; Yi, X.; Liu, S.; Sun, X. Novel InDels of GHR, GHRH, GHRHR and Their Association with Growth Traits in Seven Chinese Sheep Breeds. Animals 2020, 10, 1883. [Google Scholar] [CrossRef]
- Goodman, L.D.; Cope, H.; Nil, Z.; Ravenscroft, T.A.; Charng, W.L.; Lu, S.; Tien, A.C.; Pfundt, R.; Koolen, D.A.; Haaxma, C.A.; et al. TNPO2 variants associate with human developmental delays, neurologic deficits, and dysmorphic features and alter TNPO2 activity in Drosophila. Am. J. Hum. Genet. 2021, 108, 1669–1691. [Google Scholar] [CrossRef]
- Tan, Y.; Renqing, D.; Chun, M.; Guo, X.; Ma, X. Association of UCP2 and UCP3 gene polymorphisms and growth traits in Oula-type Tibetan sheep. J. Northwest AF Univ. Nat. Sci. Ed. 2022, 50, 9. [Google Scholar] [CrossRef]
- Cirera, S.; Clop, A.; Jacobsen, M.J.; Guerin, M.; Lesnik, P.; Jorgensen, C.B.; Fredholm, M.; Karlskov-Mortensen, P. A targeted genotyping approach enhances identification of variants in taste receptor and appetite/reward genes of potential functional importance for obesity-related porcine traits. Anim Genet. 2018, 49, 110–118. [Google Scholar] [CrossRef]
- Zhou, Z.; Han, K.; Wu, Y.; Bai, H.; Ke, Q.; Pu, F.; Wang, Y.; Xu, P. Genome-Wide Association Study of Growth and Body-Shape-Related Traits in Large Yellow Croaker (Larimichthys crocea) Using ddRAD Sequencing. Mar. Biotechnol. 2019, 21, 655–670. [Google Scholar] [CrossRef]
- Zhao, L.; Li, F.; Yuan, L.; Zhang, X.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Song, Q.; Wang, J.; et al. Expression of ovine CTNNA3 and CAP2 genes and their association with growth traits. Gene 2022, 807, 145949. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, T.; Liu, Y.; Wang, Z.; Xu, L.; Zhu, B.; Gao, X.; Zhang, L.; Gao, H.; Liu, G.E.; et al. Genome-Wide Assessment of Runs of Homozygosity in Chinese Wagyu Beef Cattle. Animals 2020, 10, 1425. [Google Scholar] [CrossRef]
- He, W.; Fang, X.; Lu, X.; Liu, Y.; Li, G.; Zhao, Z.; Li, J.; Yang, R. Function Identification of Bovine ACSF3 Gene and Its Association With Lipid Metabolism Traits in Beef Cattle. Front. Vet. Sci. 2021, 8, 766765. [Google Scholar] [CrossRef]
- Chen, G.; Cheng, X.; Shi, G.; Zou, C.; Chen, L.; Li, J.; Li, M.; Fang, C.; Li, C. Transcriptome Analysis Reveals the Effect of Long Intergenic Noncoding RNAs on Pig Muscle Growth and Fat Deposition. Biomed. Res. Int. 2019, 2019, 2951427. [Google Scholar] [CrossRef]
- Luna-Nevarez, G.; Pendleton, A.L.; Luna-Ramirez, R.I.; Limesand, S.W.; Reyna-Granados, J.R.; Luna-Nevarez, P. Genome-wide association study of a thermo-tolerance indicator in pregnant ewes exposed to an artificial heat-stressed environment. J. Biol. 2021, 101, 103095. [Google Scholar] [CrossRef]
- Zhao, P.; Zhao, F.; Hu, J.; Wang, J.; Liu, X.; Zhao, Z.; Xi, Q.; Sun, H.; Li, S.; Luo, Y. Physiology and Transcriptomics Analysis Reveal the Contribution of Lungs on High-Altitude Hypoxia Adaptation in Tibetan Sheep. Front. Physiol. 2022, 13, 885444. [Google Scholar] [CrossRef]
- Kusza, S.; Cziszter, L.T.; Ilie, D.E.; Sauer, M.; Padeanu, I.; Gavojdian, D. Kompetitive Allele Specific PCR (KASP) genotyping of 48 polymorphisms at different caprine loci in French Alpine and Saanen goat breeds and their association with milk composition. PeerJ 2018, 6, e4416. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, J.; Shi, C.; Zhu, L.; He, Q.; Tian, H.; Wu, J.; Zhao, J.; Li, C. Genome-wide analysis of the acyl-coenzyme A synthetase family and their association with the formation of goat milk flavour. Front. Genet. 2022, 13, 980463. [Google Scholar] [CrossRef]
- Armstrong, E.; Ciappesoni, G.; Iriarte, W.; Da Silva, C.; Macedo, F.; Navajas, E.A.; Brito, G.; San Julian, R.; Gimeno, D.; Postiglioni, A. Novel genetic polymorphisms associated with carcass traits in grazing Texel sheep. Meat Sci. 2018, 145, 202–208. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhao, B.; Huang, X.; Fu, X.; Liu, G.; Tian, Y.; Wu, C.; Mao, J.; Liu, J.; Gun, S.; et al. Gene network analysis reveals candidate genes related with the hair follicle development in sheep. BMC Genom. 2022, 23, 428. [Google Scholar] [CrossRef] [PubMed]
- Smolucha, G.; Gurgul, A.; Jasielczuk, I.; Kawecka, A.; Miksza-Cybulska, A. A genome-wide association study for prolificacy in three Polish sheep breeds. J. Appl. Genet. 2021, 62, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Zhang, J.; Huang, Y.; Zhao, Y.; Na, R.; Zhao, Z.; Ma, Y.; Chu, M.; Basang, W.; Zhu, Y.B.; et al. Genome-wide selection signatures analysis of litter size in Dazu black goats using single-nucleotide polymorphism. 3 Biotech 2019, 9, 336. [Google Scholar] [CrossRef]



| Trait/Number | Samples/Number | |
|---|---|---|
| Trait 1 | Trait 0 | |
| Litter size/154 | More than 2 lambs/7 | 1 to 2 lambs/147 |
| Coat colour/196 | Brown/10 | White/186 |
| Black middorsal stripe 1/196 | Present/77 | Absent/119 |
| Skin colour/205 | Pigmented/66 | Unpigmented/139 |
| Trait | CHR 1 | SNP Marker | Position | p Value | Annotated Gene |
|---|---|---|---|---|---|
| Litter size | 11 | snp54094-scaffold824-899720 | 57366312 | 6.87 × 10−7 | Near to LRRTM4 (leucine rich repeat transmembrane neuronal 4) |
| Coat colour | 12 | snp55048-scaffold842-324525 | 69164369 | 1.53 × 10−7 | LOC102187779 |
| 26 | snp11508-scaffold142-1990450 | 23574535 | 6.90 × 10−8 | SORCS3 (sortilin-related VPS10 domain-containing receptor 3) | |
| Black dorsal stripe | 18 | snp56013-scaffold873-22716 | 14190132 | 2.43 × 10−9 | TCF25 (transcription factor 25) |
| Class | ROH Number | Percent (%) | Mean Length | Genome Coverage (%) |
|---|---|---|---|---|
| ROH 2–4 Mb | 2882 | 64.82 | 2.59 | 0.30 |
| ROH 4–8 Mb | 756 | 17.00 | 5.45 | 0.16 |
| ROH 8–16 Mb | 426 | 9.58 | 10.84 | 0.18 |
| >16 Mb | 382 | 8.59 | 27.92 | 0.42 |
| Total | 4446 | 100 | 6.04 | 1.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Niu, Q.; Jiang, J.; Wang, G.; Zhou, P.; Li, J.; Chen, C.; Liu, L.; Xu, L.; Ren, H. Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers. Genes 2023, 14, 1183. https://doi.org/10.3390/genes14061183
Sun X, Niu Q, Jiang J, Wang G, Zhou P, Li J, Chen C, Liu L, Xu L, Ren H. Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers. Genes. 2023; 14(6):1183. https://doi.org/10.3390/genes14061183
Chicago/Turabian StyleSun, Xiaoyan, Qunhao Niu, Jing Jiang, Gaofu Wang, Peng Zhou, Jie Li, Cancan Chen, Liangjia Liu, Lingyang Xu, and Hangxing Ren. 2023. "Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers" Genes 14, no. 6: 1183. https://doi.org/10.3390/genes14061183
APA StyleSun, X., Niu, Q., Jiang, J., Wang, G., Zhou, P., Li, J., Chen, C., Liu, L., Xu, L., & Ren, H. (2023). Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers. Genes, 14(6), 1183. https://doi.org/10.3390/genes14061183





