The Role of Genetic and Epigenetic Regulation in Intestinal Fibrosis in Inflammatory Bowel Disease: A Descending Process or a Programmed Consequence?
Abstract
1. Introduction
2. Inflammatory Bowel Disease (IBD)
3. Intestinal Fibrosis
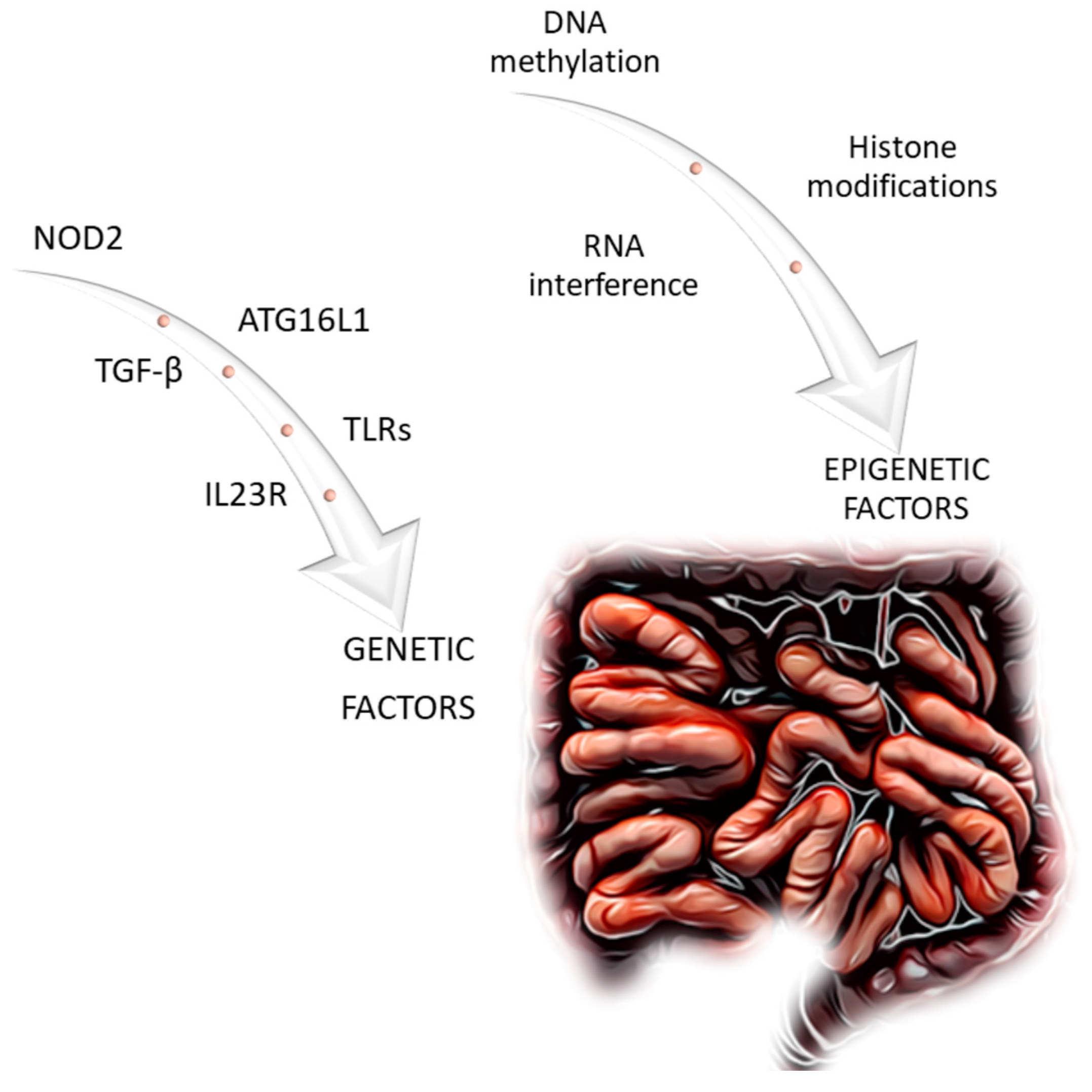
4. Genetic Factors and Mechanism of Intestinal Fibrosis in Inflammatory Bowel Disease
4.1. NOD2 (Nucleotide-Binding Oligomerization Domain-Containing 2)
4.2. TGF-β (Transforming Growth Factor β)
4.3. TLRs (Toll-like Receptors)
4.4. Il23R (Interleukin 23 Receptor)
4.5. ATG16L1 (Autophagy-Related 16-like 1)
5. Epigenetic Factors
5.1. Deoxyribonucleic Acid (DNA) Methylation
5.2. Histone Modifications
5.3. Ribonucleic Acid (RNA) Interference
6. Treatment of Intestinal Fibrosis
6.1. Potential Therapeutic Factors
6.2. Potentially Supportive Nutritional Factors
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bos, S.; Laukens, D. Metabolic modulation during intestinal fibrosis. J. Dig. Dis. 2020, 21, 319–325. [Google Scholar] [CrossRef]
- Alfredsson, J.; Wick, M.J. Mechanism of fibrosis and stricture formation in Crohn’s disease. Scand. J. Immunol. 2020, 92, e12990. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Nakase, H. The Molecular Mechanisms of Intestinal Inflammation and Fibrosis in Crohn’s Disease. Front. Physiol. 2022, 13, 845078. [Google Scholar] [CrossRef] [PubMed]
- Crespi, M.; Dulbecco, P.; De Ceglie, A.; Conio, M. Strictures in Crohn’s Disease: From Pathophysiology to Treatment. Dig. Dis. Sci. 2020, 65, 1904–1916. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, B.; Jin, T.; Ocansey, D.K.W.; Jiang, J.; Mao, F. Intestinal Fibrosis in Inflammatory Bowel Disease and the Prospects of Mesenchymal Stem Cell Therapy. Front. Immunol. 2022, 13, 835005. [Google Scholar] [CrossRef]
- Wang, J.; Lin, S.; Brown, J.M.; van Wagoner, D.; Fiocchi, C.; Rieder, F. Novel mechanisms and clinical trial endpoints in intestinal fibrosis. Immunol. Rev. 2021, 302, 211–227. [Google Scholar] [CrossRef]
- Mak, J.W.Y.; Ng, S.C. Epidemiology of fibrostenosing inflammatory bowel disease. J. Dig. Dis. 2020, 21, 332–335. [Google Scholar] [CrossRef]
- Lin, S.-N.; Mao, R.; Qian, C.; Bettenworth, D.; Wang, J.; Li, J.; Bruining, D.H.; Jairath, V.; Feagan, B.G.; Chen, M.-H.; et al. Development of antifibrotic therapy for stricturing Crohn’s disease: Lessons from randomized trials in other fibrotic diseases. Physiol. Rev. 2022, 102, 605–652. [Google Scholar] [CrossRef]
- M’koma, A.E. Inflammatory Bowel Disease: Clinical Diagnosis and Surgical Treatment-Overview. Medicina 2022, 58, 567. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Kuemmerle, J.F. Genetic and epigenetic regulation of intestinal fibrosis. United Eur. Gastroenterol. J. 2016, 4, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Mrowicki, J.; Mrowicka, M.; Majsterek, I. Czynniki środowiskowe zwiększające ryzyko aktywacji i rozwoju chorób zapalnych jelit. Postępy Biochem. 2020, 66, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Loddo, I.; Romano, C. Inflammatory Bowel Disease: Genetics, Epigenetics, and Pathogenesis. Front. Immunol. 2015, 6, 551. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Bernstein, C.N. Environmental risk factors for inflammatory bowel disease. United Eur. Gastroenterol. J. 2022, 10, 1047–1053. [Google Scholar] [CrossRef]
- El-Salhy, M.; Solomon, T.; Hausken, T.; Gilja, O.H.; Hatlebakk, J.G. Gastrointestinal neuroendocrine peptides/amines in inflammatory bowel disease. World J. Gastroenterol. 2017, 23, 5068–5085. [Google Scholar] [CrossRef] [PubMed]
- Jarmakiewicz-Czaja, S.; Zielińska, M.; Sokal, A.; Filip, R. Genetic and Epigenetic Etiology of Inflammatory Bowel Disease: An Update. Genes 2022, 13, 2388. [Google Scholar] [CrossRef]
- Miehlke, S.; Guagnozzi, D.; Zabana, Y.; Tontini, G.E.; Fiehn, A.K.; Wildt, S.; Bohr, J.; Bonderup, O.; Bouma, G.; D’Amato, M.; et al. European guidelines on microscopic colitis: United European Gastroenterology and European Microscopic Colitis Group statements and recommendations. United Eur. Gastroenterol. J. 2021, 9, 13–37. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Windsor, J.W.; Kaplan, G.G. Evolving Epidemiology of IBD. Curr. Gastroenterol. Rep. 2019, 21, 40. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, G.; Lenti, M.V.; Di Sabatino, A. Therapeutic Targeting of Intestinal Fibrosis in Crohn’s Disease. Cells 2022, 11, 429. [Google Scholar] [CrossRef]
- D’alessio, S.; Ungaro, F.; Noviello, D.; Lovisa, S.; Peyrin-Biroulet, L.; Danese, S. Revisiting fibrosis in inflammatory bowel disease: The gut thickens. Nat. Rev. Gastroenterol. Hepatol. 2021, 19, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Bettenworth, D.; Ma, C.; Parker, C.E.; Williamson, L.A.; Nelson, S.A.; van Assche, G.; Di Sabatino, A.; Bouhnik, Y.; Stidham, R.W.; et al. An expert consensus to standardise definitions, diagnosis and treatment targets for anti-fibrotic stricture therapies in Crohn’s disease. Aliment. Pharmacol. Ther. 2018, 48, 347–357. [Google Scholar] [CrossRef]
- Antar, S.A.; Ashour, N.A.; Marawan, M.E.; Al-Karmalawy, A.A. Fibrosis: Types, Effects, Markers, Mechanisms for Disease Progression, and Its Relation with Oxidative Stress, Immunity, and Inflammation. Int. J. Mol. Sci. 2023, 24, 4004. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Fiocchi, C. Mechanisms of Tissue Remodeling in Inflammatory Bowel Disease. Dig. Dis. 2013, 31, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Pakshir, P.; Hinz, B. The big five in fibrosis: Macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. 2018, 68, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef]
- Latella, G.; Rieder, F. Intestinal fibrosis. Curr. Opin. Gastroenterol. 2017, 33, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Duffield, J.S.; Lupher, M.; Thannickal, V.J.; Wynn, T.A. Host Responses in Tissue Repair and Fibrosis. Annu. Rev. Pathol. Mech. Dis. 2013, 8, 241–276. [Google Scholar] [CrossRef]
- Li, J.; Mao, R.; Kurada, S.; Wang, J.; Lin, S.; Chandra, J.; Rieder, F. Pathogenesis of fibrostenosing Crohn’s disease. Transl. Res. 2019, 209, 39–54. [Google Scholar] [CrossRef]
- Roulis, M.; Flavell, R.A. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation 2016, 92, 116–131. [Google Scholar] [CrossRef]
- Valatas, V.; Filidou, E.; Drygiannakis, I.; Kolios, G. Stromal and immune cells in gut fibrosis: The myofibroblast and the scarface. Ann. Gastroenterol. 2017, 30, 393–404. [Google Scholar] [CrossRef]
- Friedrich, M.; Pohin, M.; Powrie, F. Cytokine Networks in the Pathophysiology of Inflammatory Bowel Disease. Immunity 2019, 50, 992–1006. [Google Scholar] [CrossRef] [PubMed]
- Arpino, V.; Brock, M.; Gill, S.E. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 2015, 44, 247–254. [Google Scholar] [CrossRef]
- Vannella, K.M.; Wynn, T.A. Mechanisms of Organ Injury and Repair by Macrophages. Annu. Rev. Physiol. 2017, 79, 593–617. [Google Scholar] [CrossRef]
- Novak, E.A.; Mollen, K.P. Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell Dev. Biol. 2015, 3, 62. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, V.K.; Lebrecht, D.; Nicholson, A.G.; Wells, A.; Bhayani, H.; Gazdhar, A.; Tamm, M.; Venhoff, N.; Geiser, T.; Walker, U.A. Mitochondrial DNA mutations and respiratory chain dysfunction in idiopathic and connective tissue disease-related lung fibrosis. Sci. Rep. 2019, 9, 5500. [Google Scholar] [CrossRef]
- Latella, G.; Di Gregorio, J.; Flati, V.; Rieder, F.; Lawrance, I.C. Mechanisms of initiation and progression of intestinal fibrosis in IBD. Scand. J. Gastroenterol. 2014, 50, 53–65. [Google Scholar] [CrossRef]
- Verstockt, B.; Cleynen, I. Genetic Influences on the Development of Fibrosis in Crohn’s Disease. Front. Med. 2016, 3, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, Y.; Zhou, C.; Wu, H.; Zhao, J.; Wu, L.; Zhao, M.; Zhang, F.; Liu, H. The Role of Autophagy and Related MicroRNAs in Inflammatory Bowel Disease. Gastroenterol. Res. Pract. 2018, 2018, 7565076. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Kuemmerle, J.F. Mechanisms That Mediate the Development of Fibrosis in Patients with Crohn’s Disease. Inflamm. Bowel Dis. 2014, 20, 1250–1258. [Google Scholar] [CrossRef]
- Ueno, A.; Jijon, H.B.; Peng, R.; Sparksman, S.; Mainoli, B.; Filyk, A.; Li, Y.; Wilson, S.; Novak, K.; Panaccione, R.; et al. Association of Circulating Fibrocytes with Fibrostenotic Small Bowel Crohn’s Disease. Inflamm. Bowel Dis. 2021, 28, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Cleynen, I.; Boucher, G.; Jostins, L.; Schumm, L.P.; Zeissig, S.; Ahmad, T.; Andersen, V.; Andrews, J.M.; Annese, V.; Brand, S.; et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: A genetic association study. Lancet 2015, 387, 156–167. [Google Scholar] [CrossRef]
- Strong, L.M.; Chang, C.; Riley, J.F.; Boecker, C.A.; Flower, T.G.; Buffalo, C.Z.; Ren, X.; Stavoe, A.K.; Holzbaur, E.L.; Hurley, J.H.; et al. Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy. eLife 2021, 10, e70372. [Google Scholar] [CrossRef] [PubMed]
- Danielpour, D.; Song, K. Cross-talk between IGF-I and TGF-β signaling pathways. Cytokine Growth Factor Rev. 2006, 17, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Bettenworth, D.; Imai, J.; Inagaki, Y. Intestinal Fibrosis and Liver Fibrosis: Consequences of Chronic Inflammation or Independent Pathophysiology? Inflamm. Intest. Dis. 2016, 1, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.; Velazquez, V.M.; Brigstock, D.R. Fibrogenic Signaling Is Suppressed in Hepatic Stellate Cells through Targeting of Connective Tissue Growth Factor (CCN2) by Cellular or Exosomal MicroRNA-199a-5p. Am. J. Pathol. 2016, 186, 2921–2933. [Google Scholar] [CrossRef]
- Yun, S.-M.; Kim, S.-H.; Kim, E.-H. The Molecular Mechanism of Transforming Growth Factor-β Signaling for Intestinal Fibrosis: A Mini-Review. Front. Pharmacol. 2019, 10, 162. [Google Scholar] [CrossRef]
- Andoh, A.; Nishida, A. Molecular Basis of Intestinal Fibrosis in Inflammatory Bowel Disease. Inflamm. Intest. Dis. 2022, 7, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Stolfi, C.; Troncone, E.; Marafini, I.; Monteleone, G. Role of TGF-β and Smad7 in Gut Inflammation, Fibrosis and Cancer. Biomolecules 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Sameer, A.S.; Nissar, S. Toll-Like Receptors (TLRs): Structure, Functions, Signaling, and Role of Their Polymorphisms in Colorectal Cancer Susceptibility. BioMed Res. Int. 2021, 2021, 1157023. [Google Scholar] [CrossRef]
- Brown, M.; Hughes, K.R.; Moossavi, S.; Robins, A.; Mahida, Y.R. Toll-like receptor expression in crypt epithelial cells, putative stem cells and intestinal myofibroblasts isolated from controls and patients with inflammatory bowel disease. Clin. Exp. Immunol. 2014, 178, 28–39. [Google Scholar] [CrossRef]
- Jun, Y.K.; Kwon, S.H.; Yoon, H.T.; Park, H.; Soh, H.; Lee, H.J.; Im, J.P.; Kim, J.S.; Kim, J.W.; Koh, S.-J. Toll-like receptor 4 regulates intestinal fibrosis via cytokine expression and epithelial-mesenchymal transition. Sci. Rep. 2020, 10, 19867. [Google Scholar] [CrossRef]
- Sewell, G.W.; Kaser, A. Interleukin-23 in the Pathogenesis of Inflammatory Bowel Disease and Implications for Therapeutic Intervention. J. Crohn’s Colitis 2022, 16, ii3–ii19. [Google Scholar] [CrossRef]
- Barrett, J.C.; Hansoul, S.; Nicolae, D.L.; Cho, J.H.; Duerr, R.H.; Rioux, J.D.; Brant, S.R.; Silverberg, M.S.; Taylor, K.D.; Barmada, M.M.; et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008, 40, 955–962. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://medlineplus.gov/genetics/gene/atg16l1/#conditions (accessed on 22 April 2023).
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/gene/55054 (accessed on 23 April 2023).
- Cleynen, I.; González, J.R.; Figueroa, C.; Franke, A.; McGovern, D.; Bortlík, M.; Crusius, B.J.A.; Vecchi, M.; Artieda, M.; Szczypiorska, M.; et al. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: Results from the IBDchip European Project. Gut 2012, 62, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
- Mathur, R.; Alam, M.M.; Zhao, X.-F.; Liao, Y.; Shen, J.; Morgan, S.; Huang, T.; Lee, H.; Lee, E.; Huang, Y.; et al. Induction of autophagy in Cx3cr1+ mononuclear cells limits IL-23/IL-22 axis-mediated intestinal fibrosis. Mucosal Immunol. 2019, 12, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Kraiczy, J.; Zilbauer, M. Intestinal Epithelial Organoids as Tools to Study Epigenetics in Gut Health and Disease. Stem Cells Int. 2019, 2019, 7242415. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Modern epigenetics methods in biological research. Methods 2020, 187, 104–113. [Google Scholar] [CrossRef]
- Nacev, B.A.; Jones, K.B.; Intlekofer, A.M.; Yu, J.S.E.; Allis, C.D.; Tap, W.D.; Ladanyi, M.; Nielsen, T.O. The epigenomics of sarcoma. Nat. Rev. Cancer 2020, 20, 608–623. [Google Scholar] [CrossRef]
- Stelmaszyk, A.; Dworacka, I.M. The importance of epigenetic factors for the diagnostics and treatment of type 2. Clin. Diabetol. 2018, 7, 164–170. [Google Scholar] [CrossRef]
- Xue, T.; Qiu, X.; Liu, H.; Gan, C.; Tan, Z.; Xie, Y.; Wang, Y.; Ye, T. Epigenetic regulation in fibrosis progress. Pharmacol. Res. 2021, 173, 105910. [Google Scholar] [CrossRef] [PubMed]
- Fitz-James, M.H.; Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022, 23, 325–341. [Google Scholar] [CrossRef]
- Pawlicka, K.; Perrigue, P.; Barciszewski, J. Epigenetic control of the cellular processes. Nauka 2018, 2, 115–128. [Google Scholar]
- Marrocco, I.; Altieri, F.; Peluso, I. Measurement and Clinical Significance of Biomarkers of Oxidative Stress in Humans. Oxidative Med. Cell. Longev. 2017, 2017, 6501046. [Google Scholar] [CrossRef]
- Shen, L.; Song, C.-X.; He, C.; Zhang, Y. Mechanism and Function of Oxidative Reversal of DNA and RNA Methylation. Annu. Rev. Biochem. 2014, 83, 585–614. [Google Scholar] [CrossRef]
- Niu, Y.; DesMarais, T.L.; Tong, Z.; Yao, Y.; Costa, M. Oxidative stress alters global histone modification and DNA methylation. Free. Radic. Biol. Med. 2015, 82, 22–28. [Google Scholar] [CrossRef]
- Hannan, M.A.; Dash, R.; Sohag, A.A.M.; Haque, M.N.; Moon, I.S. Neuroprotection Against Oxidative Stress: Phytochemicals Targeting TrkB Signaling and the Nrf2-ARE Antioxidant System. Front. Mol. Neurosci. 2020, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Venkidasamy, B.; Subramanian, U.; Samynathan, R.; Shariati, M.A.; Rebezov, M.; Girish, S.; Thangavel, S.; Dhanapal, A.R.; Fedoseeva, N.; et al. Bioactive Compounds in Oxidative Stress-Mediated Diseases: Targeting the NRF2/ARE Signaling Pathway and Epigenetic Regulation. Antioxidants 2021, 10, 1859. [Google Scholar] [CrossRef]
- Law, P.-P.; Holland, M.L. DNA methylation at the crossroads of gene and environment interactions. Essays Biochem. 2019, 63, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Leroux, É.; Brosseau, C.; Angers, B.; Angers, A.; Breton, S. Méthylation De L’adn Mitochondrial. Med. Sci. 2021, 37, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Woo, V.; Alenghat, T. Epigenetic regulation by gut microbiota. Gut Microbes 2022, 14, 2022407. [Google Scholar] [CrossRef] [PubMed]
- Whyte, J.M.; Ellis, J.J.; Brown, M.A.; Kenna, T.J. Best practices in DNA methylation: Lessons from inflammatory bowel disease, psoriasis and ankylosing spondylitis. Thromb. Haemost. 2019, 21, 133. [Google Scholar] [CrossRef]
- Zouali, M. DNA methylation signatures of autoimmune diseases in human B lymphocytes. Clin. Immunol. 2020, 222, 108622. [Google Scholar] [CrossRef]
- Yang, I.V.; Schwartz, D.A. Epigenetics of idiopathic pulmonary fibrosis. Transl. Res. 2014, 165, 48–60. [Google Scholar] [CrossRef]
- Claveria-Cabello, A.; Colyn, L.; Arechederra, M.; Urman, J.M.; Berasain, C.; Avila, M.A.; Fernandez-Barrena, M.G. Epigenetics in Liver Fibrosis: Could HDACs be a Therapeutic Target? Cells 2020, 9, 2321. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.T.; Kennedy, N.; Hansen, R.; Ventham, N.; O’leary, K.R.; Drummond, H.E.; Noble, C.L.; El-Omar, E.; Russell, R.K.; Wilson, D.C.; et al. Two-stage Genome-wide Methylation Profiling in Childhood-onset Crohn’s Disease Implicates Epigenetic Alterations at the VMP1/MIR21 and HLA Loci. Inflamm. Bowel Dis. 2014, 20, 1784–1793. [Google Scholar] [CrossRef] [PubMed]
- Somineni, H.K.; Venkateswaran, S.; Kilaru, V.; Marigorta, U.M.; Mo, A.; Okou, D.T.; Kellermayer, R.; Mondal, K.; Cobb, D.; Walters, T.D.; et al. Blood-Derived DNA Methylation Signatures of Crohn’s Disease and Severity of Intestinal Inflammation. Gastroenterology 2019, 156, 2254–2265.e3. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.J.; Kraiczy, J.; Nayak, K.M.; Gasparetto, M.; Ross, A.; Lee, C.; Mak, T.N.; Koo, B.-K.; Kumar, N.; Lawley, T.; et al. DNA Methylation and Transcription Patterns in Intestinal Epithelial Cells from Pediatric Patients with Inflammatory Bowel Diseases Differentiate Disease Subtypes and Associate with Outcome. Gastroenterology 2018, 154, 585–598. [Google Scholar] [CrossRef]
- Nimmo, E.R.; Prendergast, J.G.; Aldhous, M.C.; Kennedy, N.A.; Henderson, P.; Drummond, H.E.; Ramsahoye, B.H.; Wilson, D.C.; Semple, C.A.; Satsangi, J. Genome-wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm. Bowel Dis. 2012, 18, 889–899. [Google Scholar] [CrossRef]
- McDermott, E.; Ryan, E.J.; Tosetto, M.; Gibson, D.; Burrage, J.; Keegan, D.; Byrne, K.; Crowe, E.; Sexton, G.; Malone, K.; et al. DNA Methylation Profiling in Inflammatory Bowel Disease Provides New Insights into Disease Pathogenesis. J. Crohn’s Colitis 2015, 10, 77–86. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Wu, X.; Wang, G.; Gu, G.; Ren, H.; Hong, Z.; Ren, J. Intestinal mucosa-derived DNA methylation signatures in the penetrating intestinal mucosal lesions of Crohn’s disease. Sci. Rep. 2021, 11, 9771. [Google Scholar] [CrossRef]
- Sadler, T.; Bhasin, J.M.; Xu, Y.; Barnholz-Sloan, J.; Chen, Y.; Ting, A.H.; Stylianou, E. Genome-wide analysis of DNA methylation and gene expression defines molecular characteristics of Crohn’s disease-associated fibrosis. Clin. Epigenetics 2016, 8, 30. [Google Scholar] [CrossRef]
- Fellows, R.; Varga-Weisz, P. Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk. Mol. Metab. 2019, 38, 100925. [Google Scholar] [CrossRef] [PubMed]
- Saha, S. Histone Modifications and Other Facets of Epigenetic Regulation in Trypanosomatids: Leaving Their Mark. mBio 2020, 11, e01079-20. [Google Scholar] [CrossRef]
- Demetriadou, C.; Koufaris, C.; Kirmizis, A. Histone N-α terminal modifications: Genome regulation at the tip of the tail. Epigenetics Chromatin 2020, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Qiu, T.; Wei, G.; Que, Y.; Wang, W.; Kong, Y.; Xie, T.; Chen, X. Role of Histone Post-Translational Modifications in Inflammatory Diseases. Front. Immunol. 2022, 13, 852272. [Google Scholar] [CrossRef]
- Roostaee, A.; Benoit, Y.D.; Boudjadi, S.; Beaulieu, J. Epigenetics in Intestinal Epithelial Cell Renewal. J. Cell. Physiol. 2016, 231, 2361–2367. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, S.; Wang, Z.; Huang, J.; Xu, L.; Tang, X.; Wan, Y.Y.; Li, Q.-J.; Symonds, A.L.J.; Long, H.; et al. Targeting EZH2 histone methyltransferase activity alleviates experimental intestinal inflammation. Nat. Commun. 2019, 10, 2427. [Google Scholar] [CrossRef]
- Pytlak, B.; Chraszczewska, B.; Prochorec-Sobieszek, M.; Szumera-Ciećkiewicz, A. EZH2 methyltransferase as a therapeutic target. Hematology 2019, 10, 9–18. [Google Scholar]
- Chen, Q.; Duan, X.; Xu, M.; Fan, H.; Dong, Y.; Wu, H.; Zhang, M.; Liu, Y.; Nan, Z.; Deng, S.; et al. BMSC-EVs regulate Th17 cell differentiation in UC via H3K27me3. Mol. Immunol. 2019, 118, 191–200. [Google Scholar] [CrossRef]
- Chen, P.; Zhu, H.; Mao, Y.; Zhuo, M.; Yu, Y.; Chen, M.; Zhao, Q.; Li, L.; Wu, M.; Ye, M. SETD8 involved in the progression of inflammatory bowel disease via epigenetically regulating p62 expression. J. Gastroenterol. Hepatol. 2021, 36, 2850–2863. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ding, J.; Zhang, H.; Shen, J.; Hao, Y.; Zhang, X.; Qi, W.; Luo, X.; Zhang, T.; Wang, N. Lactobacillus casei LH23 modulates the immune response and ameliorates DSS-induced colitis via suppressing JNK/p-38 signal pathways and enhancing histone H3K9 acetylation. Food Funct. 2020, 11, 5473–5485. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pan, X.; Ren, Z.; Li, B.; Liu, H.; Wu, C.; Dong, X.; Vos, P.; Pan, L.; Sun, J. Protein arginine methyltransferase 2 (PRMT2) promotes dextran sulfate sodium-induced colitis by inhibiting the SOCS3 promoter via histone H3R8 asymmetric dimethylation. Br. J. Pharmacol. 2021, 179, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Sadler, T.; Scarpa, M.; Rieder, F.; West, G.; Stylianou, E. Cytokine-induced Chromatin Modifications of the Type I Collagen α 2 Gene during Intestinal Endothelial-to-Mesenchymal Transition. Inflamm. Bowel Dis. 2013, 19, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Tzouvelekis, A.; Kaminski, N. Epigenetics in idiopathic pulmonary fibrosis. Biochem. Cell Biol. 2015, 93, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Perugorria, M.J.; Wilson, C.L.; Zeybel, M.; Walsh, M.; Amin, S.; Robinson, S.; White, S.A.; Burt, A.D.; Oakley, F.; Tsukamoto, H.; et al. Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology 2012, 56, 1129–1139. [Google Scholar] [CrossRef]
- Tsaprouni, L.G.; Ito, K.; Powell, J.J.; Adcock, I.M.; Punchard, N. Differential patterns of histone acetylation in inflammatory bowel diseases. J. Inflamm. 2011, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Tiffon, C. The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3425. [Google Scholar] [CrossRef]
- Sato, F.; Tsuchiya, S.; Meltzer, S.J.; Shimizu, K. MicroRNAs and epigenetics. FEBS J. 2011, 278, 1598–1609. [Google Scholar] [CrossRef]
- Calvo-Garrido, J.; Carilla-Latorre, S.; Escalante, R. Vacuole membrane protein 1, autophagy and much more. Autophagy 2008, 4, 835–837. [Google Scholar] [CrossRef]
- Latella, G.; Rogler, G.; Bamias, G.; Breynaert, C.; Florholmen, J.; Pellino, G.; Reif, S.; Speca, S.; Lawrance, I.C. Results of the 4th scientific workshop of the ECCO (I): Pathophysiology of intestinal fibrosis in IBD. J. Crohn’s Colitis 2014, 8, 1147–1165. [Google Scholar] [CrossRef]
- Branton, M.H.; Kopp, J.B. TGF-beta and fibrosis. Microbes Infect. 1999, 1, 1349–1365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zheng, L.; McGovern, D.P.; Hamill, A.M.; Ichikawa, R.; Kanazawa, Y.; Luu, J.; Kumagai, K.; Cilluffo, M.; Fukata, M.; et al. Myeloid ATG16L1 Facilitates Host-Bacteria Interactions in Maintaining Intestinal Homeostasis. J. Immunol. 2017, 198, 2133–2146. [Google Scholar] [CrossRef]
- Sleiman, J.; El Ouali, S.; Qazi, T.; Cohen, B.; Steele, S.R.; Baker, M.E.; Rieder, F. Prevention and Treatment of Stricturing Crohn’s Disease—Perspectives and Challenges. Expert Rev. Gastroenterol. Hepatol. 2020, 15, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Rimola, J.; Capozzi, N. Differentiation of fibrotic and inflammatory component of Crohn’s disease-associated strictures. Intest. Res. 2020, 18, 144–150. [Google Scholar] [CrossRef]
- Rieder, F.; Fiocchi, C.; Rogler, G. Mechanisms, Management, and Treatment of Fibrosis in Patients with Inflammatory Bowel Diseases. Gastroenterology 2017, 152, 340–350.e6. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.X.; Qiu, Y.; Zhuang, X.J.; Liu, F.; Wu, X.M.; Chen, M.H.; Mao, R. Intestinal stricture in Crohn’s disease: A 2020 update. J. Dig. Dis. 2021, 22, 390–398. [Google Scholar] [CrossRef]
- D’Haens, G.; Rieder, F.; Feagan, B.G.; Higgins, P.D.R.; Panes, J.; Maaser, C.; Rogler, G.; Löwenberg, M.; Van Der Voort, R.; Pinzani, M.; et al. Challenges in the Pathophysiology, Diagnosis and Management of Intestinal Fibrosis in Inflammatory Bowel Disease. Gastroenterology 2022, 162, 26–31. [Google Scholar] [CrossRef]
- Venu, V.K.P.; Alston, L.; Iftinca, M.; Tsai, Y.-C.; Stephens, M.; Warriyar KV, V.; Rehal, S.; Hudson, G.; Szczepanski, H.; von der Weid, P.-Y.; et al. Nr4A1 modulates inflammation-associated intestinal fibrosis and dampens fibrogenic signaling in myofibroblasts. Am. J. Physiol. Liver Physiol. 2021, 321, G280–G297. [Google Scholar] [CrossRef]
- Scheibe, K.; Kersten, C.; Schmied, A.; Vieth, M.; Primbs, T.; Carlé, B.; Knieling, F.; Claussen, J.; Klimowicz, A.C.; Zheng, J.; et al. Inhibiting Interleukin 36 Receptor Signaling Reduces Fibrosis in Mice with Chronic Intestinal Inflammation. Gastroenterology 2019, 156, 1082–1097.e11. [Google Scholar] [CrossRef]
- Mao, R.; Rieder, F. Cooling Down the Hot Potato: Anti-Interleukin 36 Therapy Prevents and Treats Experimental Intestinal Fibrosis. Gastroenterology 2019, 156, 871–873. [Google Scholar] [CrossRef]
- Elias, M.; Zhao, S.; Le, H.T.; Wang, J.; Neurath, M.F.; Neufert, C.; Fiocchi, C.; Rieder, F. IL-36 in chronic inflammation and fibrosis—bridging the gap? J. Clin. Investig. 2021, 131, e144336. [Google Scholar] [CrossRef]
- Amamou, A.; O’mahony, C.; Leboutte, M.; Savoye, G.; Ghosh, S.; Marion-Letellier, R. Gut Microbiota, Macrophages and Diet: An Intriguing New Triangle in Intestinal Fibrosis. Microorganisms 2022, 10, 490. [Google Scholar] [CrossRef]
- Cosin-Roger, J.; Ortiz-Masià, M.D.; Barrachina, M.D. Macrophages as an Emerging Source of Wnt Ligands: Relevance in Mucosal Integrity. Front. Immunol. 2019, 10, 02297. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, M.; Martín, R.; Torres-Maravilla, E.; Chadi, S.; González-Dávila, P.; Sokol, H.; Langella, P.; Chain, F.; Bermúdez-Humarán, L.G. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes 2020, 12, 1826748. [Google Scholar] [CrossRef] [PubMed]
- Marion-Letellier, R.; Leboutte, M.; Amamou, A.; Raman, M.; Savoye, G.; Ghosh, S. Diet in Intestinal Fibrosis: A Double-Edged Sword. Nutrients 2021, 13, 3148. [Google Scholar] [CrossRef] [PubMed]
- Tomas, J.; Mulet, C.; Saffarian, A.; Cavin, J.-B.; Ducroc, R.; Regnault, B.; Tan, C.K.; Duszka, K.; Burcelin, R.; Wahli, W.; et al. High-fat diet modifies the PPAR-γ pathway leading to disruption of microbial and physiological ecosystem in murine small intestine. Proc. Natl. Acad. Sci. USA 2016, 113, E5934–E5943. [Google Scholar] [CrossRef] [PubMed]
- Amamou, A.; Rouland, M.; Yaker, L.; Goichon, A.; Guérin, C.; Aziz, M.; Savoye, G.; Marion-Letellier, R. Dietary salt exacerbates intestinal fibrosis in chronic TNBS colitis via fibroblasts activation. Sci. Rep. 2021, 11, 15055. [Google Scholar] [CrossRef]
- Saidi, A.; Kasabova, M.; Vanderlynden, L.; Wartenberg, M.; Kara-Ali, G.H.; Marc, D.; Lecaille, F.; Lalmanach, G. Curcumin inhibits the TGF-β1-dependent differentiation of lung fibroblasts via PPARγ-driven upregulation of cathepsins B and L. Sci. Rep. 2019, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Ji, Y.-Y.; Dai, Y.-C.; Wen, X.-L.; Wu, S.-C. Network pharmacology and molecular docking reveal zedoary turmeric-trisomes in Inflammatory bowel disease with intestinal fibrosis. World J. Clin. Cases 2022, 10, 7674–7685. [Google Scholar] [CrossRef]
- Tao, Q.; Wang, B.; Zheng, Y.; Jiang, X.; Pan, Z.; Ren, J. Vitamin D Prevents the Intestinal Fibrosis Via Induction of Vitamin D Receptor and Inhibition of Transforming Growth Factor-Beta1/Smad3 Pathway. Dig. Dis. Sci. 2014, 60, 868–875. [Google Scholar] [CrossRef]
- Agista, A.Z.; Rusbana, T.B.; Islam, J.; Ohsaki, Y.; Sultana, H.; Hirakawa, R.; Watanabe, K.; Nochi, T.; Ardiansyah; Budijanto, S.; et al. Fermented Rice Bran Supplementation Prevents the Development of Intestinal Fibrosis Due to DSS-Induced Inflammation in Mice. Nutrients 2021, 13, 1869. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Choi, J.W.; Jhun, J.; Kwon, J.Y.; Lee, B.-I.; Yang, C.W.; Park, S.-H.; Cho, M.-L. Lactobacillus acidophilus Improves Intestinal Inflammation in an Acute Colitis Mouse Model by Regulation of Th17 and Treg Cell Balance and Fibrosis Development. J. Med. Food 2018, 21, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.F.F.A.; Sampaio-Maia, B.; Araujo, R.; Nascimento, D.S.; Ferreira-Gomes, J.; Pestana, M.; Azevedo, M.J.; Alencastre, I.S. Gut Microbiome and Organ Fibrosis. Nutrients 2022, 14, 352. [Google Scholar] [CrossRef] [PubMed]
- Jarmakiewicz-Czaja, S.; Ferenc, K.; Filip, R. Antioxidants as Protection against Reactive Oxidative Stress in Inflammatory Bowel Disease. Metabolites 2023, 13, 573. [Google Scholar] [CrossRef] [PubMed]
| Genetic and Epigenetic Factors | Site of Action | Role in Fibrosis |
|---|---|---|
| NOD2 (nucleotide-binding oligomerization domain-containing 2) |
|
|
| TGF-β (transforming growth factor β) |
|
|
| TLRs (toll-like receptors) |
|
|
| Il23R (interleukin 23 receptor) |
|
|
| ATG16L1 (autophagy-related 16-like 1) |
|
|
| DNA (deoxyribonucleic acid) methylation |
|
|
| Histone modifications |
|
|
| RNA (ribonucleic acid) interference |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarmakiewicz-Czaja, S.; Sokal, A.; Ferenc, K.; Motyka, E.; Helma, K.; Filip, R. The Role of Genetic and Epigenetic Regulation in Intestinal Fibrosis in Inflammatory Bowel Disease: A Descending Process or a Programmed Consequence? Genes 2023, 14, 1167. https://doi.org/10.3390/genes14061167
Jarmakiewicz-Czaja S, Sokal A, Ferenc K, Motyka E, Helma K, Filip R. The Role of Genetic and Epigenetic Regulation in Intestinal Fibrosis in Inflammatory Bowel Disease: A Descending Process or a Programmed Consequence? Genes. 2023; 14(6):1167. https://doi.org/10.3390/genes14061167
Chicago/Turabian StyleJarmakiewicz-Czaja, Sara, Aneta Sokal, Katarzyna Ferenc, Elżbieta Motyka, Kacper Helma, and Rafał Filip. 2023. "The Role of Genetic and Epigenetic Regulation in Intestinal Fibrosis in Inflammatory Bowel Disease: A Descending Process or a Programmed Consequence?" Genes 14, no. 6: 1167. https://doi.org/10.3390/genes14061167
APA StyleJarmakiewicz-Czaja, S., Sokal, A., Ferenc, K., Motyka, E., Helma, K., & Filip, R. (2023). The Role of Genetic and Epigenetic Regulation in Intestinal Fibrosis in Inflammatory Bowel Disease: A Descending Process or a Programmed Consequence? Genes, 14(6), 1167. https://doi.org/10.3390/genes14061167






