High Level of GMFG Correlated to Poor Clinical Outcome and Promoted Cell Migration and Invasion through EMT Pathway in Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
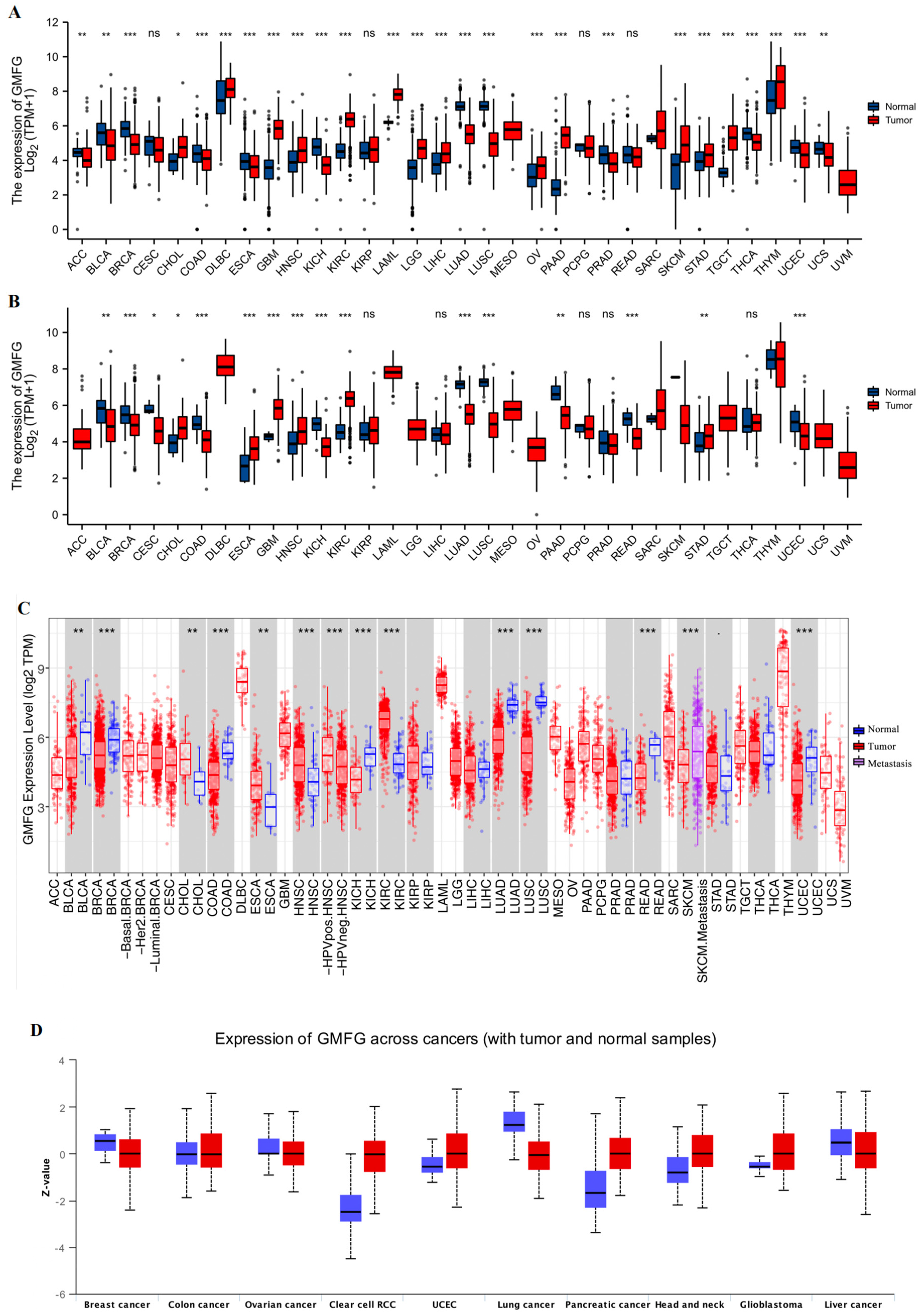
2.2. GMFG Expression Profiles in Human Cancers
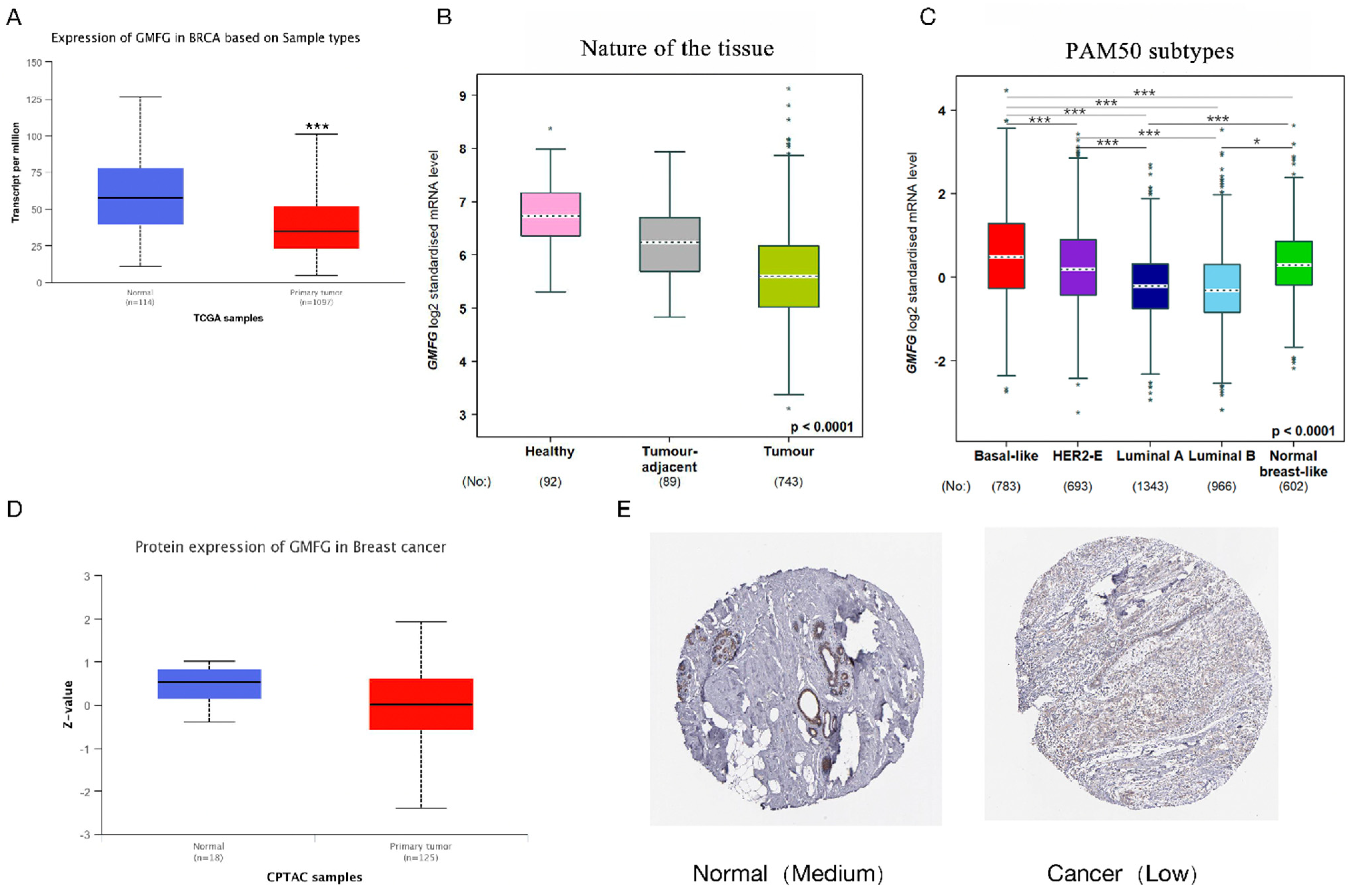
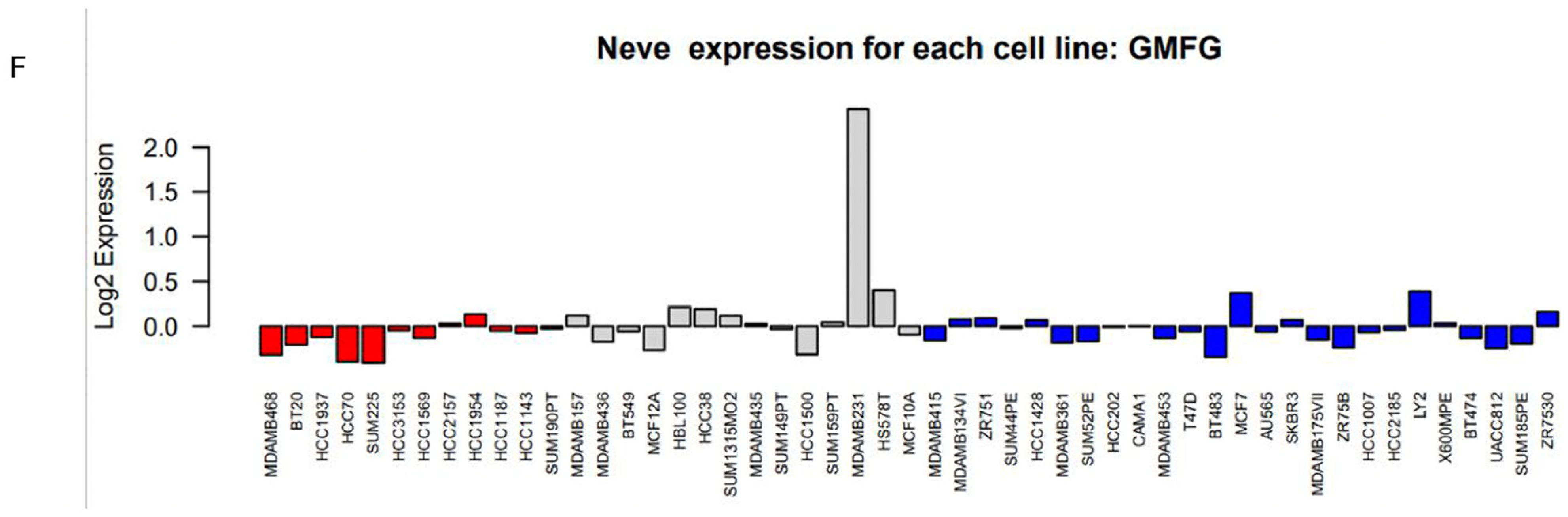
2.3. GMFG Expression Profiles in Breast Cancer
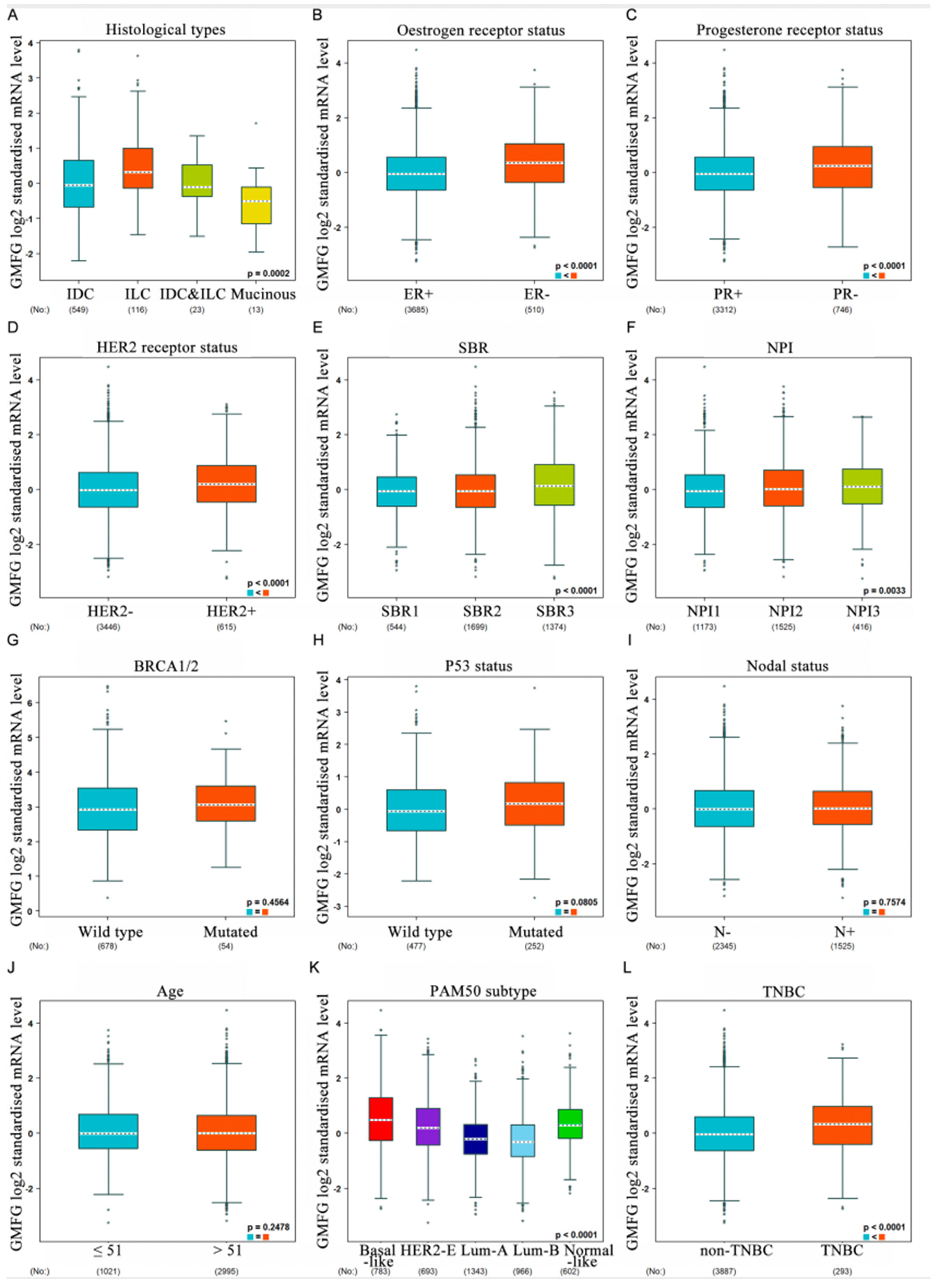
2.4. The Relationship between GMFG Expression and Breast Cancer Clinicopathological Features
2.5. Function Enrichment Analysis
2.6. Patients, Breast Cancer Tissues, and Breast Cancer Cell Lines
2.7. siRNA and Transfection
2.8. Immunoblotting
2.9. Wound Healing Assay
2.10. Transwell Migration and Invasion Assay
2.11. Immunohistochemistry
2.12. Statistical Analysis
3. Results
3.1. GMFG Is Differentially Expressed in Human Cancers
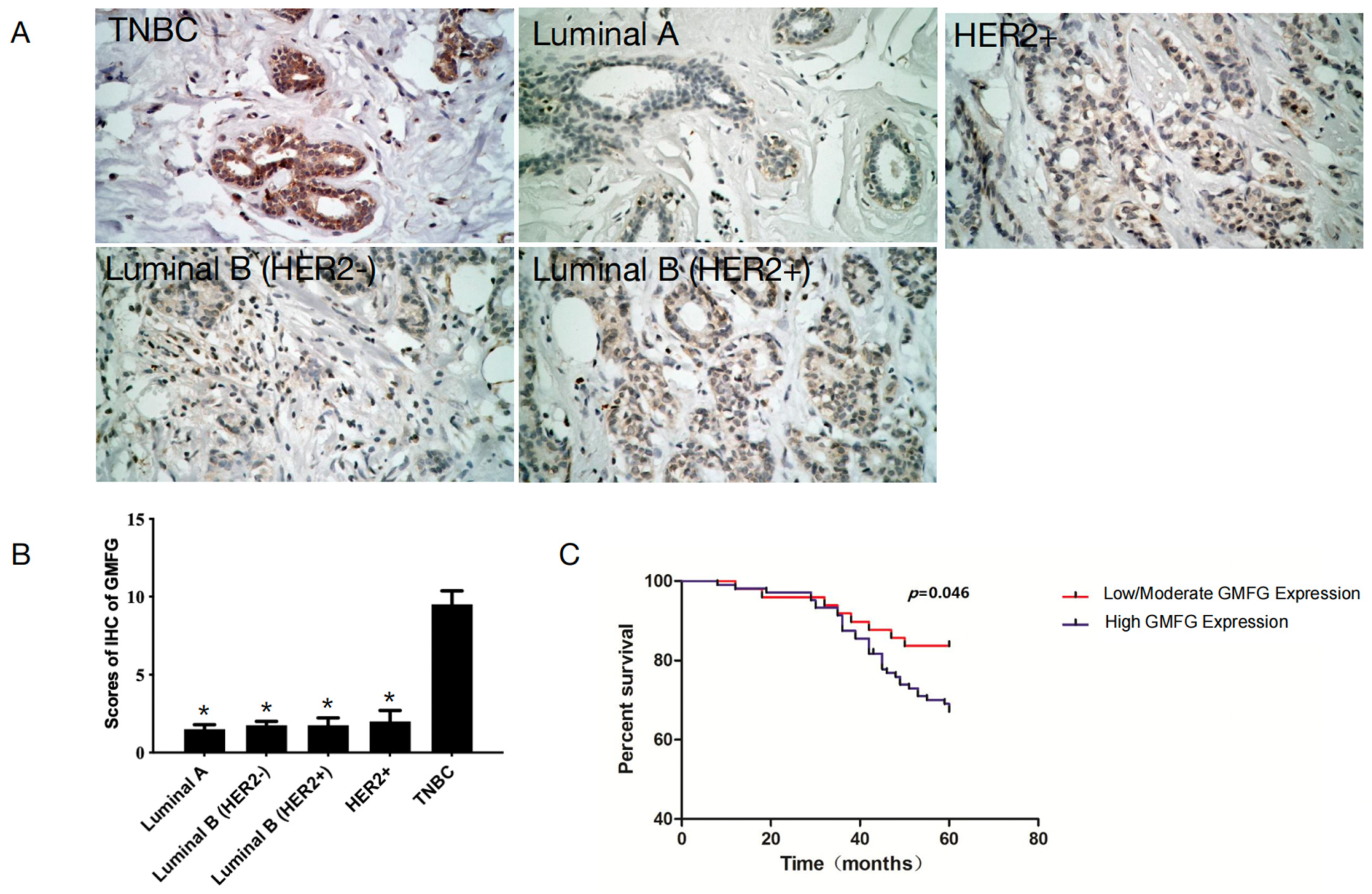
3.2. GMFG Is Differentially Expressed in Different Types of Breast Cancer and Significantly So in the Basal-like Subtype
3.3. Correlation between GMFG Expression and Clinicopathological Parameters of Breast Cancer Patients
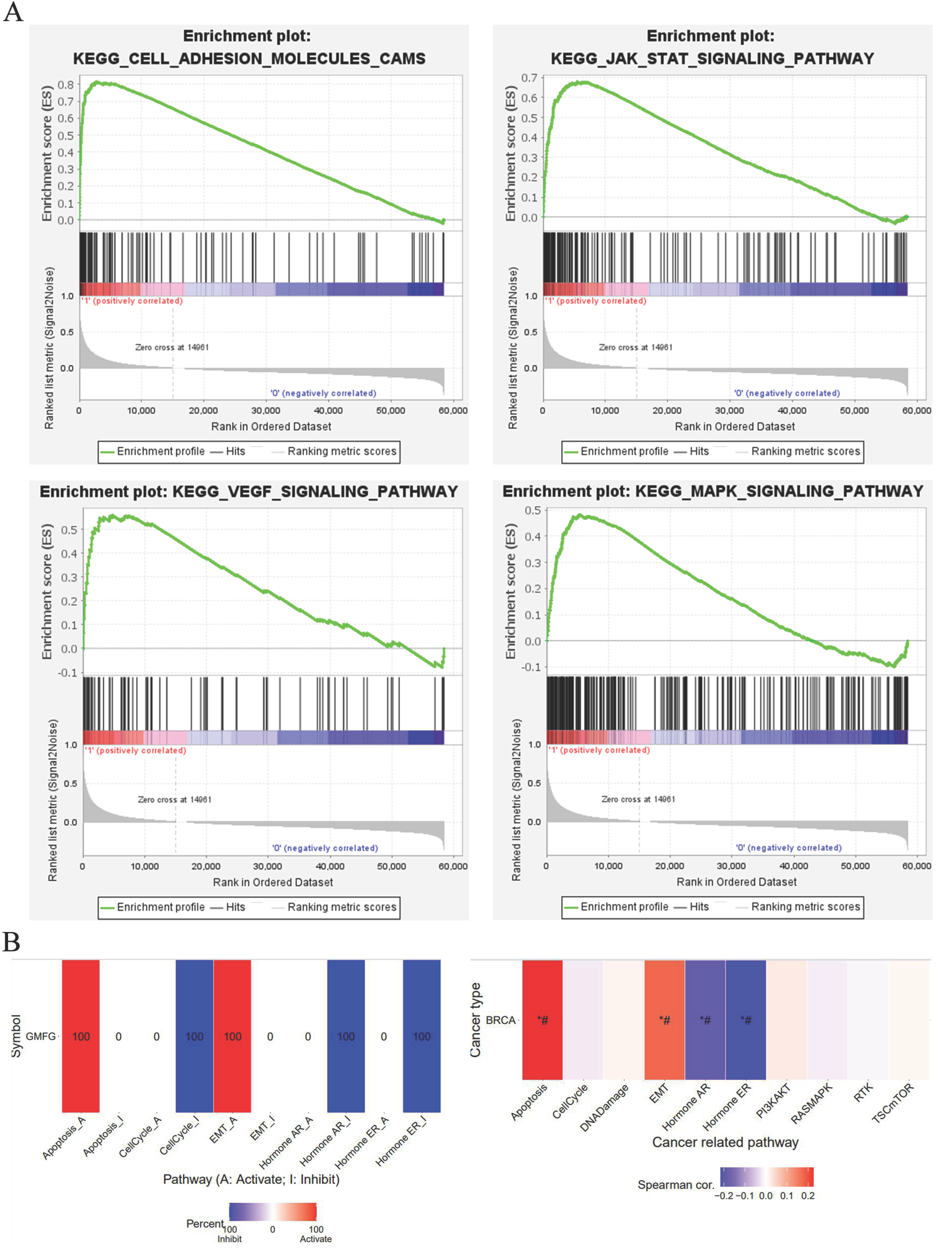
3.4. Function Enrichment Analysis
3.5. GMFG Is Significantly Upregulated in TNBC Samples and Is Related to Poor Prognosis for TNBC Patients
3.6. GMFG Promotes TNBC Cell Migration and Invasion by Regulating the TGF-β-Mediated EMT Signaling Pathway
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J. Strategies for subtypes--dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.S.; Chanana, P.; Jhamb, S. Biomarkers in triple negative breast cancer: A review. World J. Clin. Oncol. 2015, 6, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.L.; Sweeney, M.O.; Eskin, J.A. GMF as an Actin Network Remodeling Factor. Trends Cell Biol. 2018, 28, 749–760. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, L.; Liotta, L.A.; Wan, H.H.; Rodgers, G.P. Glia maturation factor gamma (GMFG): A cytokine-responsive protein during hematopoietic lineage development and its functional genomics analysis. Genom. Proteom. Bioinform. 2006, 4, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, M.; Aoyama, M.; Sobue, K.; Yamamoto, N.; Morishima, T.; Moriyama, A.; Katsuya, H.; Asai, K. Sensitive immunoassays for human and rat GMFB and GMFG, tissue distribution and age-related changes. Biochim. Biophys. Acta 2004, 1670, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Aerbajinai, W.; Lee, K.; Chin, K.; Rodgers, G.P. Glia maturation factor-gamma negatively modulates TLR4 signaling by facilitating TLR4 endocytic trafficking in macrophages. J. Immunol. 2013, 190, 6093–6103. [Google Scholar] [CrossRef]
- Aerbajinai, W.; Liu, L.; Chin, K.; Zhu, J.; Parent, C.A.; Rodgers, G.P. Glia maturation factor-gamma mediates neutrophil chemotaxis. J. Leukoc. Biol. 2011, 90, 529–538. [Google Scholar] [CrossRef]
- Aerbajinai, W.; Liu, L.; Zhu, J.; Kumkhaek, C.; Chin, K.; Rodgers, G.P. Glia Maturation Factor-gamma Regulates Monocyte Migration through Modulation of beta1-Integrin. J. Biol. Chem. 2016, 291, 8549–8564. [Google Scholar] [CrossRef]
- Lippert, D.N.; Wilkins, J.A. Glia maturation factor gamma regulates the migration and adherence of human T lymphocytes. BMC Immunol. 2012, 13, 21. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Z.; Chang, H.; Mu, X.; Deng, W.; Yuan, Z.; Yao, F.; Liu, Y.; Mai, R.; Wu, B. Expression of glia maturation factor gamma is associated with colorectal cancer metastasis and its downregulation suppresses colorectal cancer cell migration and invasion in vitro. Oncol. Rep. 2017, 37, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Zuo, P.; Ma, Y.; Huang, Y.; Ye, F.; Wang, P.; Wang, X.; Zhou, C.; Lu, W.; Kong, B.; Xie, X. High GMFG expression correlates with poor prognosis and promotes cell migration and invasion in epithelial ovarian cancer. Gynecol. Oncol. 2014, 132, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Jezequel, P.; Gouraud, W.; Azzouz, F.B.; Basseville, A.; Juin, P.P.; Lasla, H.; Campone, M. Interest of the bc-GenExMiner web tool in oncology. Bull. Cancer 2021, 108, 1057–1064. [Google Scholar] [CrossRef]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarró, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef]
- Hao, X.P.; Pretlow, T.G.; Rao, J.S.; Pretlow, T.P. Beta-catenin expression is altered in human colonic aberrant crypt foci. Cancer Res. 2001, 61, 8085–8088. [Google Scholar]
- Zhao, Y.; Zhao, Y.; Zhang, M.; Zhao, J.; Ma, X.; Huang, T.; Pang, H.; Li, J.; Song, J. Inhibition of TLR4 Signalling-Induced Inflammation Attenuates Secondary Injury after Diffuse Axonal Injury in Rats. Mediat. Inflamm. 2016, 2016, 4706915. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.; Tate, C.R.; Segar, H.C.; Burks, H.E.; Phamduy, T.B.; Hoang, V.; Elliott, S.; Gilliam, D.; Pounder, F.N.; Anbalagan, M.; et al. Suppression of triple-negative breast cancer metastasis by pan-DAC inhibitor panobinostat via inhibition of ZEB family of EMT master regulators. Breast Cancer Res. Treat. 2014, 145, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.D.; Parvani, J.G.; Schiemann, W.P. The relevance of the TGF-beta Paradox to EMT-MET programs. Cancer Lett. 2013, 341, 30–40. [Google Scholar] [CrossRef]
- Ikeda, K.; Kundu, R.K.; Ikeda, S.; Kobara, M.; Matsubara, H.; Quertermous, T. Glia maturation factor-gamma is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ. Res. 2006, 99, 424–433.R. [Google Scholar] [CrossRef] [PubMed]
- Parvani, J.G.; Gujrati, M.D.; Mack, M.A.; Schiemann, W.P.; Lu, Z.R. Silencing beta3 Integrin by Targeted ECO/siRNA Nanoparticles Inhibits EMT and Metastasis of Triple-Negative Breast Cancer. Cancer Res. 2015, 75, 2316–2325. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.O.; Fang, Y.; Li, Z.; Chen, Z.; Xiang, J. Role of cellular cytoskeleton in epithelial-mesenchymal transition process during cancer progression. Biomed. Rep. 2015, 3, 603–610. [Google Scholar] [CrossRef]
- Yu, W.; Kanaan, Y.; Bae, Y.K.; Gabrielson, E. Chromosomal changes in aggressive breast cancers with basal-like features. Cancer Genet. Cytogenet. 2009, 193, 29–37. [Google Scholar] [CrossRef]
- Kuuselo, R.; Savinainen, K.; Azorsa, D.O.; Basu, G.D.; Karhu, R.; Tuzmen, S.; Mousses, S.; Kallioniemi, A. Intersex-like (IXL) is a cell survival regulator in pancreatic cancer with 19q13 amplification. Cancer Res. 2007, 67, 1943–1949. [Google Scholar] [CrossRef]

| Clinicopathological Characteristics | GMFG | p Value | |
|---|---|---|---|
| Low/Moderate (%) | High (%) | ||
| Age (year) | |||
| <50 | 23 (30.26) | 53 (69.74) | 0.642 |
| ≥50 | 26 (33.77) | 51 (66.23) | |
| Menopause | |||
| Premenopausal | 29 (38.67) | 46 (61.33) | 0.084 |
| Postmenopausal | 20 (25.64) | 58 (74.36) | |
| Tumor size (cm) | |||
| T1 | 21 (45.65) | 25 (54.35) | 0.120 |
| T2 | 16 (26.23) | 45 (73.77) | |
| T3 | 7 (29.17) | 17 (70.83) | |
| T4 | 5 (22.73) | 17 (77.27) | |
| Histological grade | |||
| G1 | 19 (48.72) | 20 (51.28) | 0.033 * |
| G2 | 18 (27.69) | 47 (72.31) | |
| G3 | 12 (24.49) | 37 (75.51) | |
| Axillary lymph node metastasis | |||
| Negative | 32 (40.00) | 48 (60.00) | 0.027 * |
| Positive | 17 (23.29) | 56 (76.71) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Wei, X.; Li, J.; Diao, Y.; Shan, C.; Li, W.; Zhang, S.; Wu, F. High Level of GMFG Correlated to Poor Clinical Outcome and Promoted Cell Migration and Invasion through EMT Pathway in Triple-Negative Breast Cancer. Genes 2023, 14, 1157. https://doi.org/10.3390/genes14061157
Zhao Y, Wei X, Li J, Diao Y, Shan C, Li W, Zhang S, Wu F. High Level of GMFG Correlated to Poor Clinical Outcome and Promoted Cell Migration and Invasion through EMT Pathway in Triple-Negative Breast Cancer. Genes. 2023; 14(6):1157. https://doi.org/10.3390/genes14061157
Chicago/Turabian StyleZhao, Yonglin, Xing Wei, Jia Li, Yan Diao, Changyou Shan, Weimiao Li, Shuqun Zhang, and Fei Wu. 2023. "High Level of GMFG Correlated to Poor Clinical Outcome and Promoted Cell Migration and Invasion through EMT Pathway in Triple-Negative Breast Cancer" Genes 14, no. 6: 1157. https://doi.org/10.3390/genes14061157
APA StyleZhao, Y., Wei, X., Li, J., Diao, Y., Shan, C., Li, W., Zhang, S., & Wu, F. (2023). High Level of GMFG Correlated to Poor Clinical Outcome and Promoted Cell Migration and Invasion through EMT Pathway in Triple-Negative Breast Cancer. Genes, 14(6), 1157. https://doi.org/10.3390/genes14061157







