Development of Multiplex PCR and Melt–Curve Analysis for the Molecular Identification of Four Species of the Mullidae Family, Available in the Market
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Morphological Identification, and DNA Extraction
2.2. Sanger Sequencing and Primer Design
2.3. Multiplex PCR Reaction
2.4. Real-Time PCR Reaction and Melt–Curve Analysis
3. Results
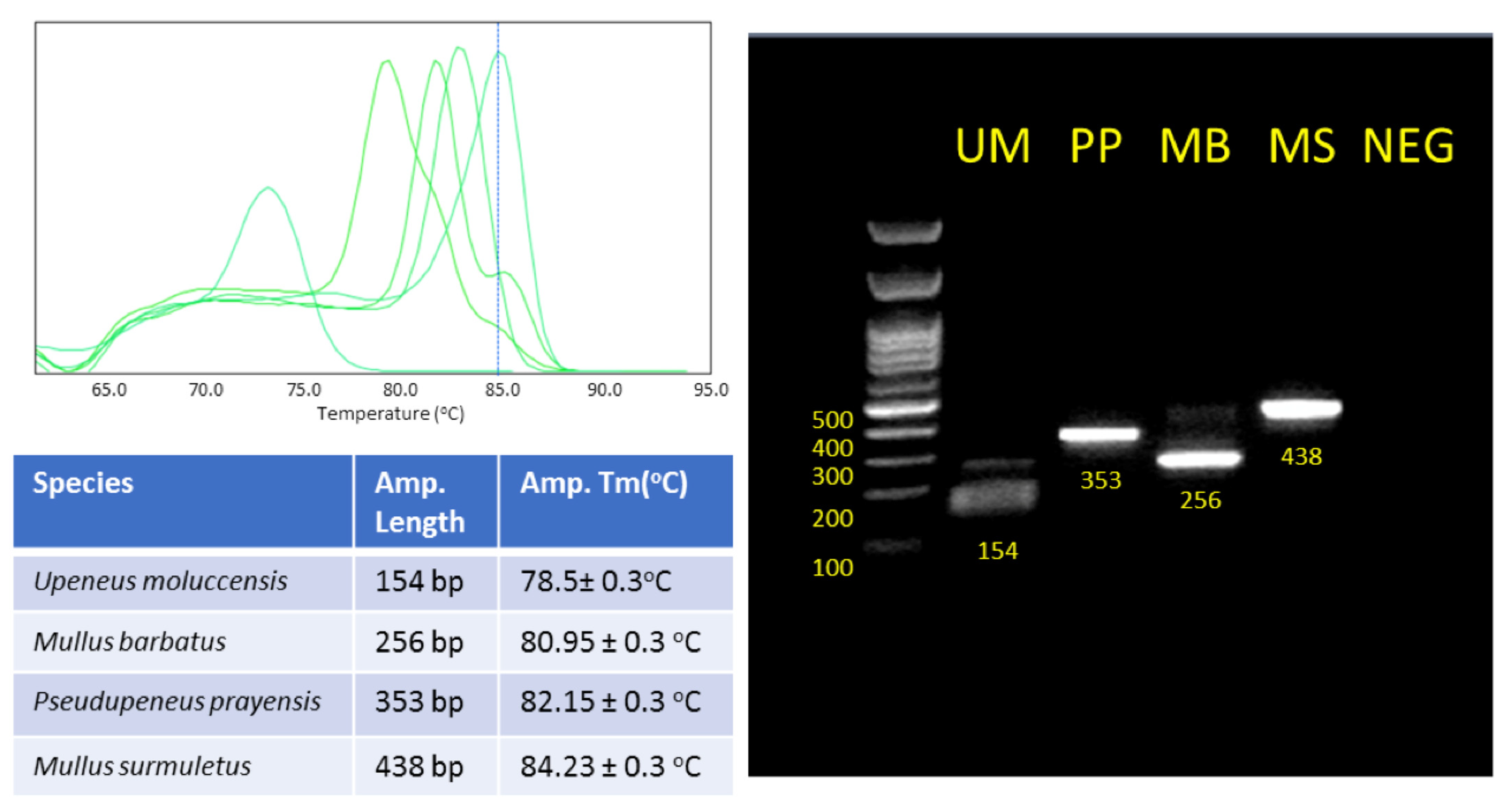
3.1. Species-Specific Primer Design and Optimization of Multiplex PCR
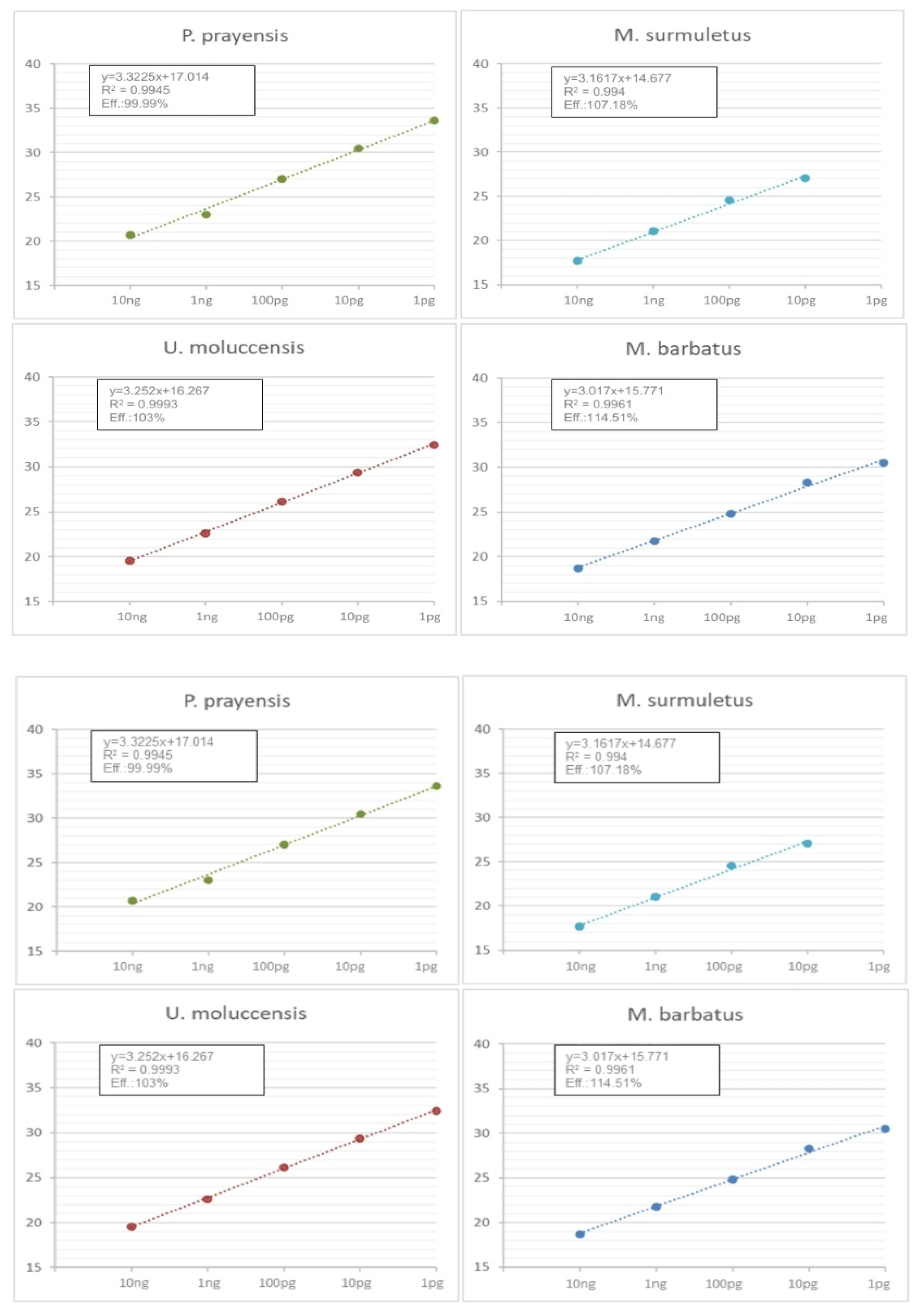
3.2. Optimization of Multiplex Real-Time PCR of CO1
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kotsanopoulos, K.V.; Exadactylos, A.; Gkafas, G.A.; Martsikalis, P.V.; Parlapani, F.F.; Boziaris, I.S.; Arvanitoyannis, I.S. The use of molecular markers in the verification of fish and seafood authenticity and the detection of adulteration. Compr. Rev. Food Sci. Food Saf. 2021, 20, 1584–1654. [Google Scholar] [CrossRef] [PubMed]
- Kotsanopoulos, K.; Martsikalis, P.V.; Gkafas, G.A.; Exadactylos, A. The use of various statistical methods for authenticity and detection of adulteration in fish and seafood. Crit. Rev. Food Sci. Nutr. 2022, 2, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R.S.; Morrissey, M.T. DNA-based methods for the identification of commercial fish and seafood species. Compr. Rev. Food Sci. Food Saf. 2008, 7, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Čapla, J.; Zajác, P.; Čurlej, J.; Belej, Ľ.; Kročko, M.; Bobko, M.; Benešová, L.; Jakabová, S.; Vlčko, T. Procedures for the identification and detection of adulteration of fish and meat products. Potravin. Slovak J. Food Sci. 2020, 14, 978–994. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- ELSTAT 2020. Quantity of Catch by Species of Catch, Category of Fishery and Type of Fishing Gear (January 2020). Available online: https://www.statistics.gr (accessed on 20 January 2023).
- Arvanitoyannis, I.S.; Krystallis, A.; Panagiotaki, P.; Theodorou, A.J. A marketing survey on Greek consumers’ attitudes towards fish. Aquac. Intern. 2004, 12, 259–279. [Google Scholar] [CrossRef]
- Nelson, J.S. Fishes of the World, 3rd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 1994; 600 p. [Google Scholar]
- Golani, D.; Öztürk, B.; Başusta, N. The Fishes of the Eastern Mediterranean; Turkish Marine Research Foundation: Istanbul, Turkey, 2006; pp. 173–174. [Google Scholar]
- Nicolaidou, A.; Alongi, G.; Aydogan, O.; Catra, M.; Cavas, L.; Cevik, C.; Vazquez-Luis, M.; Filiz, H. New Mediterranean Biodiversity Records (June 2012). Mediterr. Mar. Sci. 2012, 13, 162. [Google Scholar] [CrossRef]
- Apostolidis, A.P.; Mamuris, Z.; Triantaphyllidis, C. Phylogenetic relationships among four species of Mullidae (Perciformes) inferred from DNA sequences of mitochondrial cytochrome b and 16S rRNA genes. Biochem. Syst. Ecol. 2001, 29, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Mamuris, Z.; Apostolidis, P.; Panagiotaki, P.; Theodorou, A.J.; Triantaphyllidis, C. Morphological variation between red mullet populations in Greece. J. Fish Biol. 1998, 52, 107–117. [Google Scholar] [CrossRef]
- Calvo, J.H.; Osta, R.; Zaragoza, P. Quantitative PCR detection of pork in raw and heated ground beef and Păte. J Agric. Food Chem. 2002, 50, 5265–5267. [Google Scholar] [CrossRef] [PubMed]
- von Bargen, C.; Brockmeyer, J.; Humpf, H. Meat authentication: A new HPLC—MS/MS based method for the fast and sensitive detection of horse and pork in highly processed food. J. Agric. Food Chem. 2014, 62, 9428–9435. [Google Scholar] [CrossRef] [PubMed]
- Handy, S.M.; Deeds, J.R.; Ivanova, N.V.; Hebert, P.D.; Hanner, R.H.; Ormos, A.; Weigt, L.A.; Moore, M.M.; Yancy, H.F. A single-laboratory validated method for the generation of DNA barcodes for the identification of fish for regulatory compliance. J. AOAC Int. 2011, 94, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Pääbo, S.; Villablanca, F.X.; Wilson, A.C. 1989. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Fernandez, C.; Ardura, A.; Masiá, P.; Rodriguez, N.; Voces, L.; Fernandez-Raigoso, M.; Roca, A.; Machado-Schiaffino, G.; Dopico, E.; Garcia-Vazquez, E. Fraud in highly appreciated fish detected from DNA in Europe may undermine the Development Goal of sustainable fishing in Africa. Sci. Rep. 2021, 11, 11423. [Google Scholar] [CrossRef] [PubMed]
- Cutarelli, A.; Amoroso, M.G.; De Roma, A.; Girardi, S.; Galiero, G.; Guarino, A.; Corrado, F. Italian market fish species identification and commercial frauds revealing by DNA sequencing. Food Control 2014, 37, 46–50. [Google Scholar] [CrossRef]
- Mamuris, Z.; Stamatis, C.; Moutou, K.A.; Apostolidis, A.P.; Triantaphyllidis, C. RFLP Analysis of Mitochondrial DNA to Evaluate Genetic Variation in Striped Red Mullet (Mullus surmuletus L.) and Red Mullet (Mullus barbatus L.) Populations. Mar. Biotechnol. 2001, 3, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, D.A.; Emel’yanova, N.G.; Thuan, L.T.B.; Ha, V.T. Reproduction and initial development of manybar goatfish Parupeneus multifasciatus (Mullidae). J. Ichthyol. 2011, 51, 604–617. [Google Scholar] [CrossRef]
- Deidun, A.; Zava, B.; Insacco, G.; Foka, M. First record of the Por’s goatfish Upeneus pori (Actinopterygii: Perciformes: Mullidae) from Italian waters (western Ionian Sea). Acta Ichthyol. Et Piscat. 2018, 48, 93–97. [Google Scholar] [CrossRef]
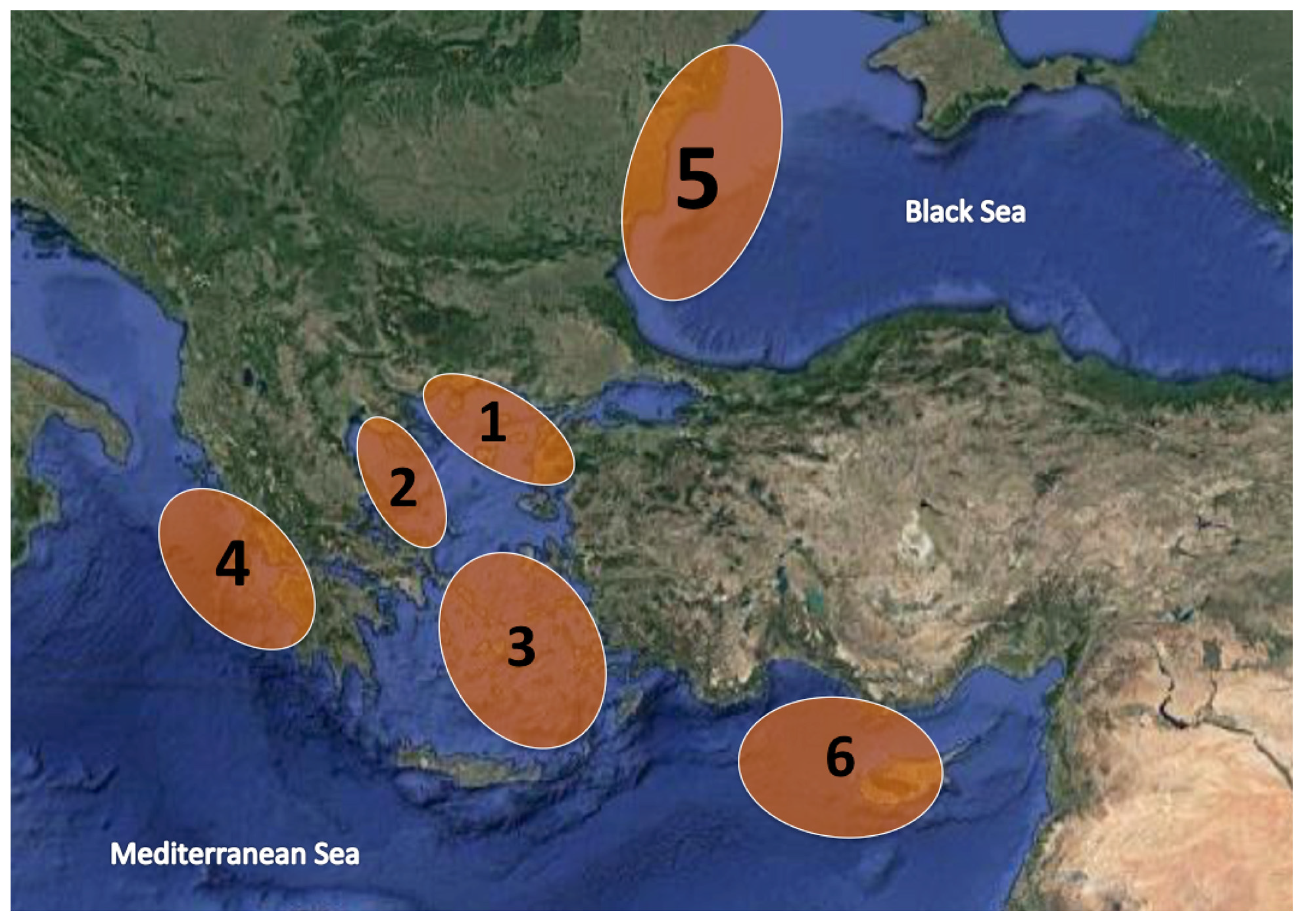


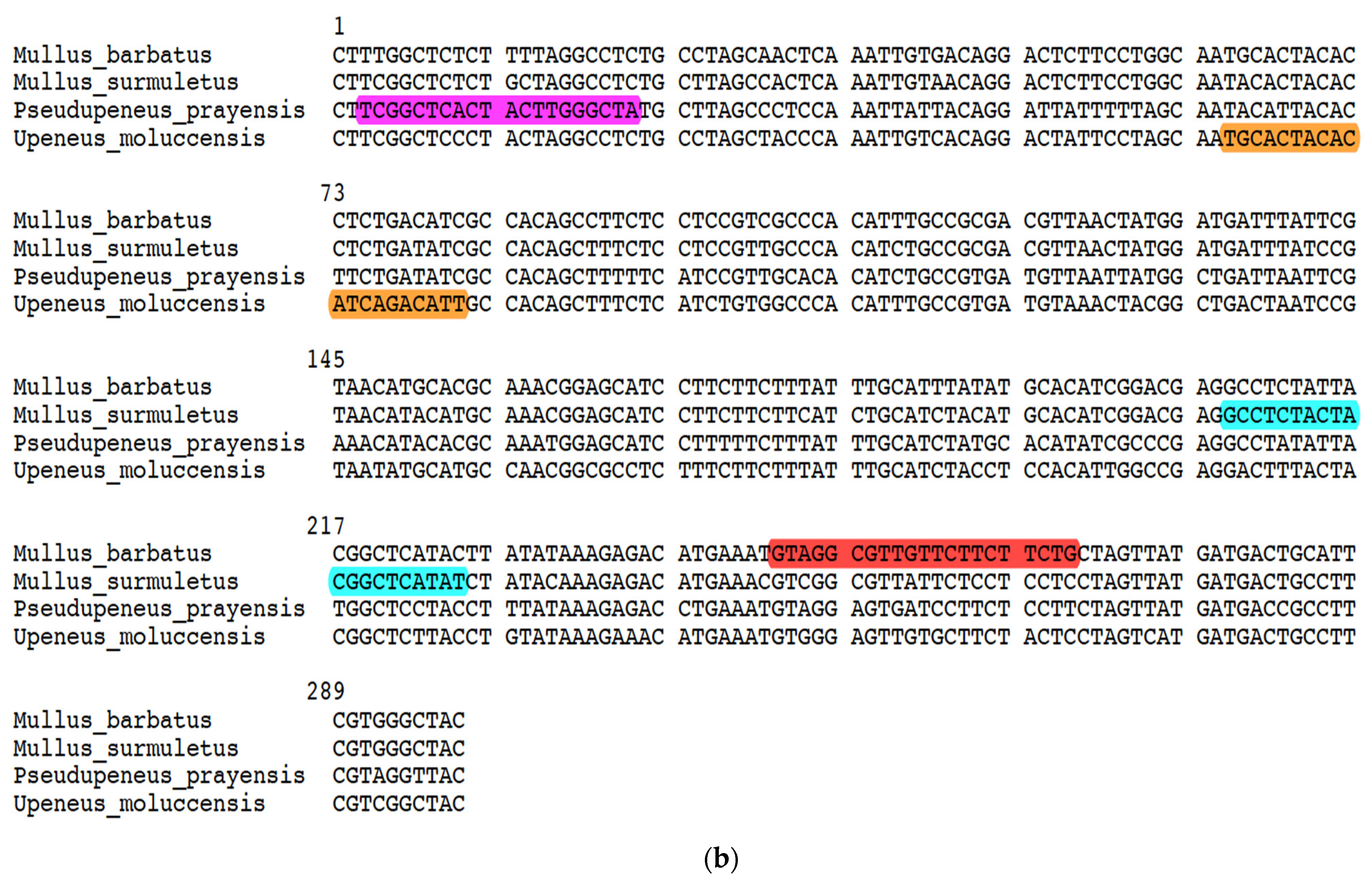
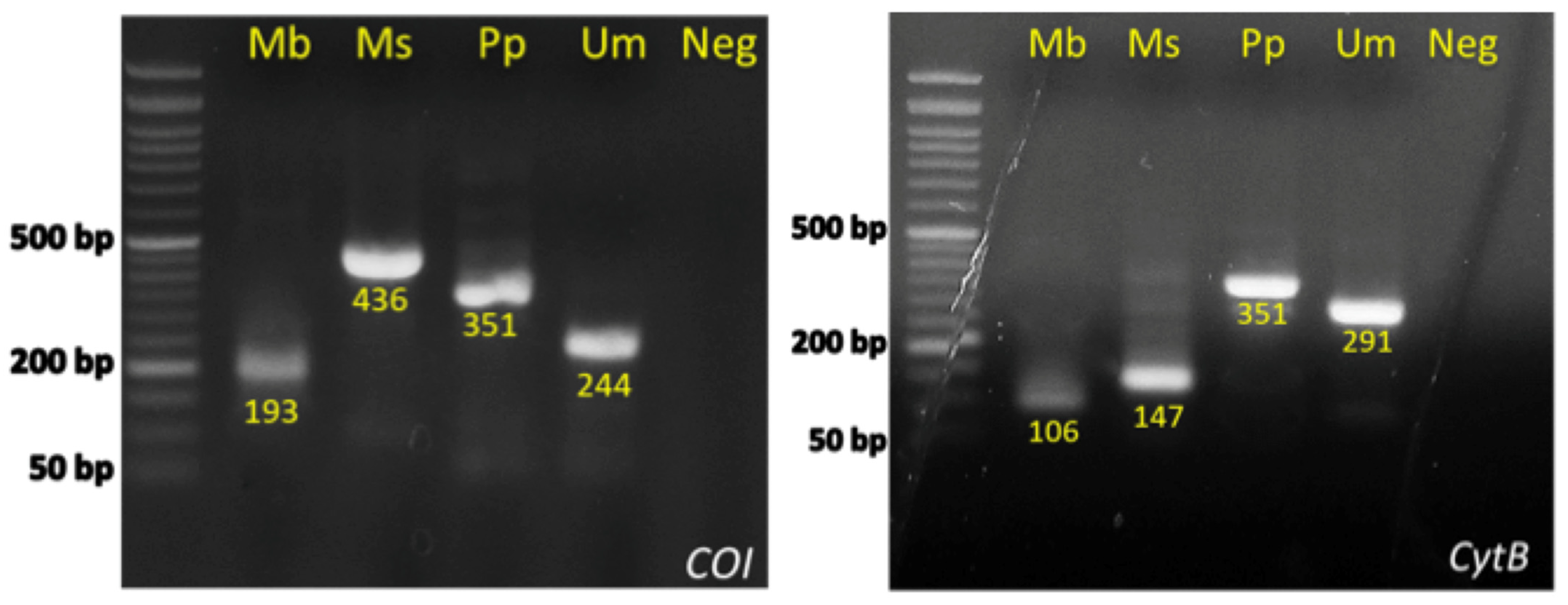
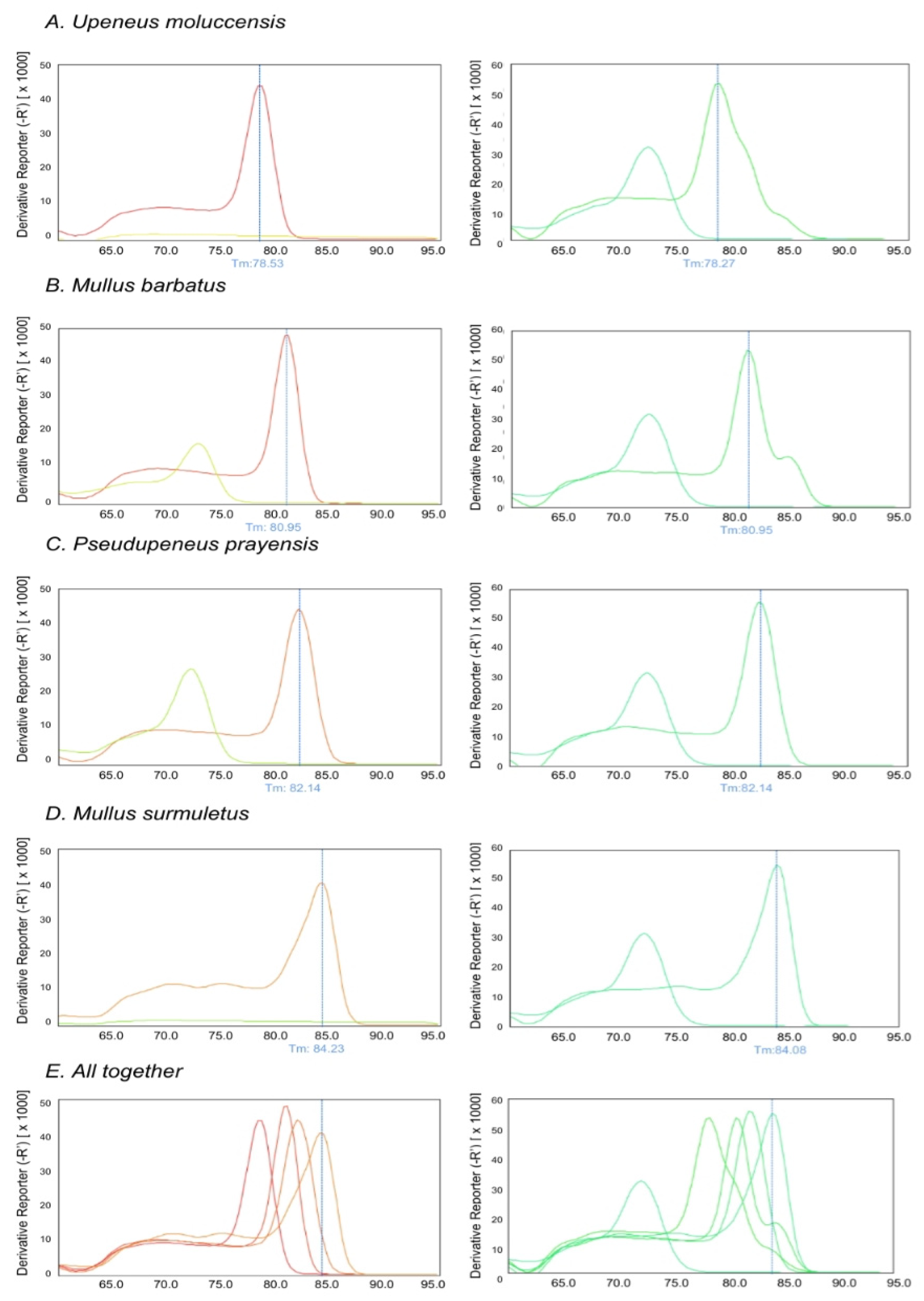
| Marine Area/Species | Mullus surmuletus | Mullus barbatus | Upeneus moluccensis | Pseudupeneus prayensis |
|---|---|---|---|---|
| Northeast Aegean (1) | 34 | 30 | - | - |
| Northwest Aegean (2) | 31 | 30 | - | - |
| Central Aegean (3) | 12 | 18 | - | - |
| Ionian Sea (4) | 21 | 5 | - | - |
| Black Sea (5) | - | 62 | - | - |
| Cyprus (6) | 20 | 20 | 15 | |
| Atlantic (FAO34) (7) | - | - | - | 20 |
| Total | 118 | 165 | 15 | 20 |
| Name | Sequence | Tm | Amplicon Length | Gene Target | Targeted Organism |
|---|---|---|---|---|---|
| Primer Mix for Multiplex CO1 | |||||
| COIUniF | AAGCCTYCTYATTCGTGC | 60.18 | - | CO1 | Universal |
| COIMbR | TTCGGGGGAAAGCCATATCG | 62.54 | 193 | CO1 | Mullus barbatus |
| COIUmR | GGCAAGCAGTAGCAGGAAAGAA | 63.62 | 244 | CO1 | Upeneus moluccensis |
| COIPpR | AAGAGAAAAAATAGTTAAGTCAACG | 56.59 | 351 | CO1 | Pseudupeneus prayensis |
| COIMsR | ATTGTGAAATTGCTGGAGGC | 59.58 | 436 | CO1 | Mullus surmuletus |
| Primer Mix for Multiplex CYTB | |||||
| Cyt bUni/H15149 | AAACTGCAGCCCCTCAGAATGATATTTGTCCTCA | 71.01 | - | CYTB | Universal |
| CyMbF | GTAGGCGTTRTTCTTCTTCTG | 59.41 | 106 | CYTB | Mullus barbatus |
| CyPpF | TCGGCTCACTACTTGGGCTA | 63.01 | 351 | CYTB | Pseudupeneus prayensis |
| CyMsF | GCCTCTACTACGGCTCATAT | 58.64 | 147 | CYTB | Mullus surmuletus |
| CyUmF | TGCACTACACATCAGACATT | 57.32 | 291 | CYTB | Upeneus moluccensis |
| Primer Mix for Melt–Curve real-time PCR CO1 | |||||
| 2COIUniF | AAGCCTYCTYATTCGTGC | 60.18 | - | CO1 | Universal |
| 2COIUmR | TGGGATAAGTCAGTTACCAAAT | 54.7 | 154 | CO1 | Upeneus moluccensis |
| 2COIMbR | AACGCCTGAAGAGGCTAGTAGA | 60.3 | 256 | CO1 | Mullus barbatus |
| 2COIPpR | AAGAGAAAAAATAGTTAAGTCAACG | 54.8 | 353 | CO1 | Pseudupeneus prayensis |
| 2COIMsR | ATTGTGAAATTGCTGGAGGC | 55.3 | 438 | CO1 | Mullus surmuletus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giantsis, I.A.; Tokamani, M.; Triantaphyllidis, G.; Tzatzani, S.; Chatzinikolaou, E.; Toros, A.; Bouchorikou, A.; Chatzoglou, E.; Miliou, H.; Sarantopoulou, J.; et al. Development of Multiplex PCR and Melt–Curve Analysis for the Molecular Identification of Four Species of the Mullidae Family, Available in the Market. Genes 2023, 14, 960. https://doi.org/10.3390/genes14050960
Giantsis IA, Tokamani M, Triantaphyllidis G, Tzatzani S, Chatzinikolaou E, Toros A, Bouchorikou A, Chatzoglou E, Miliou H, Sarantopoulou J, et al. Development of Multiplex PCR and Melt–Curve Analysis for the Molecular Identification of Four Species of the Mullidae Family, Available in the Market. Genes. 2023; 14(5):960. https://doi.org/10.3390/genes14050960
Chicago/Turabian StyleGiantsis, Ioannis A., Maria Tokamani, George Triantaphyllidis, Stella Tzatzani, Emmanuella Chatzinikolaou, Athanasios Toros, Anastasia Bouchorikou, Evanthia Chatzoglou, Helen Miliou, Joanne Sarantopoulou, and et al. 2023. "Development of Multiplex PCR and Melt–Curve Analysis for the Molecular Identification of Four Species of the Mullidae Family, Available in the Market" Genes 14, no. 5: 960. https://doi.org/10.3390/genes14050960
APA StyleGiantsis, I. A., Tokamani, M., Triantaphyllidis, G., Tzatzani, S., Chatzinikolaou, E., Toros, A., Bouchorikou, A., Chatzoglou, E., Miliou, H., Sarantopoulou, J., Gkafas, G. A., Exadactylos, A., Sandaltzopoulos, R., & Apostolidis, A. P. (2023). Development of Multiplex PCR and Melt–Curve Analysis for the Molecular Identification of Four Species of the Mullidae Family, Available in the Market. Genes, 14(5), 960. https://doi.org/10.3390/genes14050960












