The Complete Mitochondrial Genome and Phylogenetic Analyses of To Chicken in Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Polymerase Chain Reaction (PCR) Amplifications of the mtDNA Sequences
2.3. Sequencing and Structure Analysis
2.4. Phylogenetic Analysis
3. Results
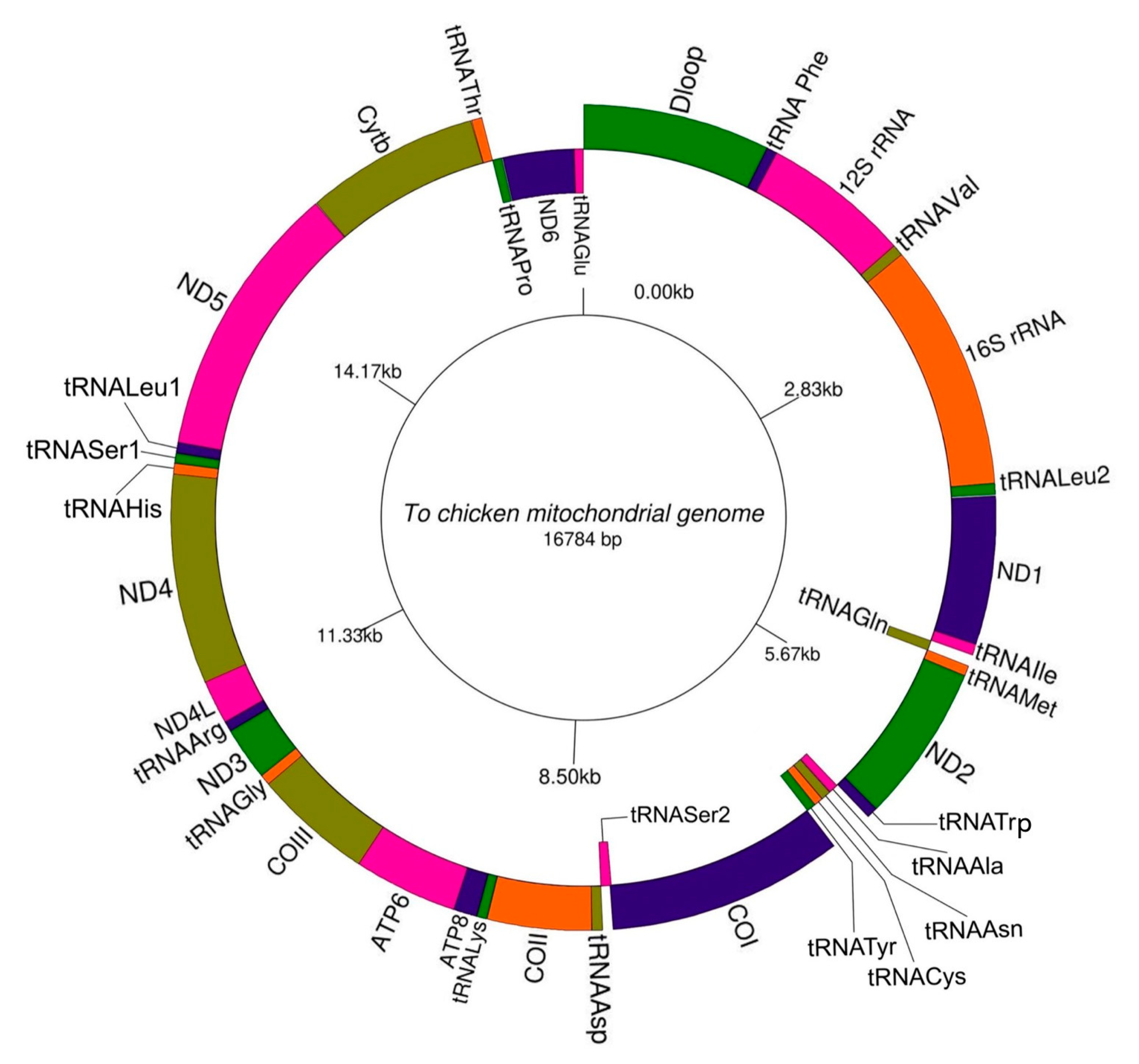
3.1. Structure of Mitochondrial Genome of To Chicken
3.2. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phuong, T.L.; Xuan, K.D.; Szalay, I. Traditions and local use of native Vietnamese chicken breeds in sustainable rural farming. World’s Poult. Sci. J. 2015, 71, 385–396. [Google Scholar] [CrossRef]
- Pham, T.C.; Nguyen, T.Q.; Pham, N.H.; Ho, T.X.; Tran, P.V.; Tran, H.T.; Nguyen, H.T.T.; Ma, D.T.; Pham, B.H.; Le, B.T. Selection of nuclear herd of to chickens. J. Anim. Sci. Technol. 2018, 85, 46–54. [Google Scholar]
- Do, S.Q.; Nguyen, L.T.P.; Nguyen, T.H.; Nguyen, T.Q. Genomic characterization of three Vietnamese indigenous chicken varieties using mitochondrial D-loop sequences. Can. J. Anim. Sci. 2019, 99, 833–839. [Google Scholar] [CrossRef]
- Arif, I.A.; Khan, H.A.; Bahkali, A.H.; Al Homaidan, A.A.; Al Farhan, A.H.; Al Sadoon, M.; Shobrak, M. DNA marker technology for wildlife conservation. Saudi J. Biol. Sci. 2011, 18, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Nabholz, B.; Glémin, S.; Hurst, G.D.D. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Mol. Ecol. 2009, 18, 4541–4550. [Google Scholar] [CrossRef]
- Liu, Y.P.; Wu, G.S.; Yao, Y.G.; Miao, Y.W.; Luikart, G.; Baig, M.; Beja-Pereira, A.; Ding, Z.L.; Palanichamy, M.G.; Zhang, Y.P. Multiple maternal origins of chickens: Out of the Asian jungles. Mol. Phylogenetics Evol. 2006, 38, 12–19. [Google Scholar] [CrossRef]
- Oka, T.; Ino, Y.; Nomura, K.; Kawashima, S.; Kuwayama, T.; Hanada, H.; Amano, T.; Takada, M.; Takahata, N.; Hayashi, Y.; et al. Analysis of mtDNA sequences shows Japanese native chickens have multiple origins. Anim. Genet. 2007, 38, 287–293. [Google Scholar] [CrossRef]
- Osman, S.A.M.; Yonezawa, T.; Nishibori, M. Origin and genetic diversity of Egyptian native chickens based on complete sequence of mitochondrial DNA D-loop region. Poult. Sci. 2016, 95, 1248–1256. [Google Scholar] [CrossRef]
- Kawabe, K.; Worawut, R.; Taura, S.; Shimogiri, T.; Nishida, T.; Okamoto, S. Genetic diversity of mtDNA D-loop polymor-phisms in Laotian native fowl populations. Asian-Australas. J. Anim. Sci. 2014, 27, 19. [Google Scholar] [CrossRef]
- Ulfah, M.; Perwitasari, D.; Jakaria, J.; Muladno, M.; Farajallah, A. Multiple maternal origins of Indonesian crowing chickens revealed by mitochondrial DNA analysis. Mitochondrial DNA Part A 2017, 28, 254–262. [Google Scholar] [CrossRef]
- Teinlek, P.; Siripattarapravat, K.; Tirawattanawanich, C. Genetic diversity analysis of Thai indigenous chickens based on complete sequences of mitochondrial DNA D-loop region. Asian-Australas. J. Anim. Sci. 2018, 31, 804. [Google Scholar] [CrossRef]
- Godinez, C.J.P.; Dadios, P.J.D.; Espina, D.M.; Matsunaga, M.; Nishibori, M. Population Genetic Structure and Contribution of Philippine Chickens to the Pacific Chicken Diversity Inferred From Mitochondrial DNA. Front. Genet. 2021, 12, 698401. [Google Scholar] [CrossRef]
- Cuc, N.T.K.; Simianer, H.; Groeneveld, L.F.; Weigend, S. Multiple maternal lineages of Vietnamese local chickens inferred by mitochondrial DNA D-loop sequences. Asian-Australas. J. Anim. Sci. 2011, 24, 155–161. [Google Scholar] [CrossRef]
- Nguyen, T.; Duc, N.; Khoa, D.; Tuong, N.; Reyer, H.; Wimmers, K.; Thuy, D.; Thuy, N. Genetic diversity of Vietnamese native chicken breeds based on mitochondrial DNA D-loop sequence. J. Anim. Plant Sci. 2022, 32, 653–662. [Google Scholar]
- Berthouly-Salazar, C.; Rognon, X.; Nhu Van, T.; Gély, M.; Vu Chi, C.; Tixier-Boichard, M.; Bed’Hom, B.; Bruneau, N.; Verrier, E.; Maillard, J.C. Vietnamese chickens: A gate towards Asian genetic diversity. BMC Genet. 2010, 11, 53. [Google Scholar] [CrossRef]
- Miao, Y.W.; Peng, M.S.; Wu, G.S.; Ouyang, Y.N.; Yang, Z.Y.; Yu, N.; Liang, J.P.; Pianchou, G.; Beja-Pereira, A.; Mitra, B.; et al. Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity 2013, 110, 277–282. [Google Scholar] [CrossRef]
- Powell, A.F.L.A.; Barker, K.F.; Lanyon, S.M. Empirical evaluation of partitioning schemes for phylogenetic analyses of mitogenomic data: An avian case study. Mol. Phylogenetics Evol. 2013, 66, 69–79. [Google Scholar] [CrossRef]
- Achilli, A.; Bonfiglio, S.; Olivieri, A.; Malusa, A.; Pala, M.; Kashani, B.H.; Perego, U.A.; Ajmone-Marsan, P.; Liotta, L.; Semino, O. The multifaceted origin of taurine cattle reflected by the mitochondrial genome. PLoS ONE 2009, 4, e5753. [Google Scholar] [CrossRef]
- Achilli, A.; Olivieri, A.; Pellecchia, M.; Uboldi, C.; Colli, L.; Al-Zahery, N.; Accetturo, M.; Pala, M.; Kashani, B.H.; Perego, U.A.; et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr. Biol. 2008, 18, R157–R158. [Google Scholar] [CrossRef]
- Bonfiglio, S.; Achilli, A.; Olivieri, A.; Negrini, R.; Colli, L.; Liotta, L.; Ajmone-Marsan, P.; Torroni, A.; Ferretti, L. The enigmatic origin of bovine mtDNA haplogroup R: Sporadic interbreeding or an independent event of Bos primigenius domestication in Italy? PLoS ONE 2010, 5, e15760. [Google Scholar] [CrossRef]
- Pang, J.F.; Kluetsch, C.; Zou, X.J.; Zhang, A.B.; Luo, L.Y.; Angleby, H.; Ardalan, A.; Ekström, C.; Sköllermo, A.; Lundeberg, J.; et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol. Biol. Evol. 2009, 26, 2849–2864. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Kashani, B.H.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.S.; Yao, Y.G.; Qu, K.X.; Ding, Z.L.; Li, H.; Palanichamy, M.G.; Duan, Z.Y.; Li, N.; Chen, Y.S.; Zhang, Y.P. Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biol. 2007, 8, R245. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.G.; Zhao, C.J.; Li, J.Y.; Wu, C.X. Sequencing and alignment of mitochondrial genomes of Tibetan chicken and two lowland chicken breeds. Sci. China Ser. C Life Sci. 2008, 51, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Xia, X. DAMBE6: New tools for microbial genomics, phylogenetics, and molecular evolution. J. Hered. 2017, 108, 431–437. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Rao, Y.S.; Chai, X.W.; Wang, Z.F.; Nie, Q.H.; Zhang, X.Q. Impact of GC content on gene expression pattern in chicken. Genet. Sel. Evol. 2013, 45, 9. [Google Scholar] [CrossRef]
- Desjardins, P.; Morais, R. Sequence and gene organization of the chicken mitochondrial genome: A novel gene order in higher vertebrates. J. Mol. Biol. 1990, 212, 599–634. [Google Scholar] [CrossRef]
- Clayton, D.A. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 1991, 7, 453–478. [Google Scholar] [CrossRef]
- Huang, X.H.; Zhong, F.S.; Li, W.N.; Chen, J.B.; Zhang, A.X.; Yao, Q.F. Complete mitochondrial genome of the Wuhua three-yellow chicken (Gallus gallus domesticus). Mitochondrial DNA Part A 2016, 27, 1311–1312. [Google Scholar] [CrossRef]
- Gu, J.; Li, S. Complete mitochondrial genome of the Longsheng Feng chicken (Gallus gallus). Mitochondrial DNA Part B 2020, 5, 2911–2912. [Google Scholar] [CrossRef]
- Gu, J.; Li, S. The complete mitochondrial genome of the Luhua chicken (Gallus gallus). Mitochondrial DNA Part B 2020, 5, 2832–2834. [Google Scholar] [CrossRef]
- Gu, J.; Li, S. Next-generation sequencing of the complete mitochondrial genome of the Piao chicken (Gallus gallus). Mitochondrial DNA Part B 2020, 5, 2870–2871. [Google Scholar] [CrossRef]
- Liu, L.L.; Xie, H.B.; Yu, Q.F.; He, S.P.; He, J.H. Determination and analysis of the complete mitochondrial genome sequence of Taoyuan chicken. Mitochondrial DNA Part A 2016, 27, 371–372. [Google Scholar] [CrossRef]
- Jin, S.; Zang, H.; He, P.; Jiang, T.; Pan, S.; Geng, Z. Complete mitochondrial genome and phylogenetic analysis of Huangshan Black chicken (Gallus gallus). Mitochondrial DNA Part B 2021, 6, 243–244. [Google Scholar] [CrossRef]
- Kanakachari, M.; Chatterjee, R.; Rajkumar, U.; Haunshi, S.; Reddy, M.; Bhattacharya, T. Indian Red Jungle fowl depicts close genetic relationship with Indian native chicken breeds as evidenced through whole mitochondrial genome intersection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Nishibori, M.; Shimogiri, T.; Hayashi, T.; Yasue, H. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim. Genet. 2005, 36, 367–375. [Google Scholar] [CrossRef]
- Lawal, R.A.; Hanotte, O. Domestic chicken diversity: Origin, distribution, and adaptation. Anim. Genet. 2021, 52, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.; Larson, G.; Gunnarsson, U.; Bed’Hom, B.; Tixier-Boichard, M.; Strömstedt, L.; Wright, D.; Jungerius, A.; Vereijken, A.; Randi, E.; et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008, 4, e1000010. [Google Scholar] [CrossRef] [PubMed]
- DeSalle, R.; Schierwater, B.; Hadrys, H. MtDNA: The small workhorse of evolutionary studies. Front. Bioscie. 2017, 22, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.H.; Wu, H.; Fujihara, T.; Fang, S.G. Which genetic marker for which conservation genetics issue? Electrophoresis 2004, 25, 2165–2176. [Google Scholar] [CrossRef]
- Mueller, J.P.; Rischkowsky, B.; Haile, A.; Philipsson, J.; Mwai, O.; Besbes, B.; Valle Zárate, A.; Tibbo, M.; Mirkena, T.; Duguma, G. Community—Based livestock breeding programmes: Essentials and examples. J. Anim. Breed. Gen. 2015, 132, 155–168. [Google Scholar] [CrossRef]


| Primer | PCR Primer Sequences | Ta * °C | Expected PCR Product Length (bp) | |
|---|---|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | |||
| 1 | CTCGCCCTACTTGCCTTCC | TGCCTGATACCTGCTCCTTT | 60 | 925 |
| 2 | CACTGAAGATGCCAAGATGGTA | CCTTGACCTGTCTTATTAGCGA | 63 | 748 |
| 3 | TGCCAGCACAGCCTACATA | GAGACGGGTTCGCTCAAAT | 58 | 750 |
| 4 | TAGCAAGAACAACCAAGCAAAGTG | CCATTCATACAAGTCTCAATTTACGG | 63 | 781 |
| 5 | GCAAACCAAAGACCCGACTG | GTGTTCATAGATAGAAACCGACCTG | 60 | 656 |
| 6 | CGACAAGGAGGTTTACGAC | GAAGGTTTGTTAGGGTGGG | 62 | 389 |
| 7 | TTAACAGTCCTACGTGATCTGAGT | GAGTGCAATGGGAAATAATTCT | 59 | 1077 |
| 8 | TAACCACCGTCCTATTCCTG | ATCAGGCGTTGGTTATGCT | 63 | 699 |
| 9 | CGAGCGATTGAAGCCACTA | GCAAGTCGGAGGTAGAAGAAT | 59 | 740 |
| 10 | GGCTTCATGCCAAAATGACT | AGAATGGAGGAAACACCTGCTA | 55.5 | 1109 |
| 11 | CCTACTAGCCTCATCTACCGTAG | GCGTCTGGGTAATCTGAGTATC | 59 | 1105 |
| 12 | ACAGGCTTTACCCTACACCCA | GGTTAAGATGACAGTAGTGAGGATCA | 60 | 1172 |
| 13 | GCAATCCCTGGACGACTAAATCA | ATGGGCTTGGGTCAACTATGTG | 62 | 1122 |
| 14 | ACCAATAATACCATCAATCTCC | CGCTTAGTAGAAAGGATAGTGAG | 59 | 1034 |
| 15 | TTTGCCTCCTACGACTAATCAA | GCTGTATATTGTGGTGTTAGTTCATAT | 54 | 1008 |
| 16 | TCATTCGCCCTTGGACCTAT | TTGGGGTGGGTGAGTTTGAT | 58 | 682 |
| 17 | CATTCGCCCTTGGACCTATC | GATGGAAGAGTGCCTCGTTGG | 63 | 1388 |
| 18 | CCTAAAATCCCTCATTGCCTAC | TATGTTATTTGCGATGGTTAGTG | 60 | 1157 |
| 19 | GAAAGCATTGCCACCCACTGA | TGATTGCTGGGGTTCGTGTG | 60 | 1141 |
| 20 | CCACCTCCTGCCTAACCATT | TCGTCCGATGTGAAGGAAGATA | 60 | 1020 |
| 21 | AACGTACAATACGGCTGAC | AGGTTTGAGTCCTCCTTTT | 61 | 1022 |
| 22 | CCCCACAATCGGAACACTA | GGTCTAACCAAGCGGGAATA | 60 | 735 |
| 23 | GATTAGACGCCACAGCTAAA | TTCGTGAAAAGTGAGAAAGTTC | 63 | 721 |
| No. | Breed/Common Name | GenBank Accession No. | Geographic Location |
|---|---|---|---|
| 1 | Red jungle fowl (G. g. bankiva) | AP003323 | Indonesia |
| 2 | Red jungle fowl (G. g. gallus) | AP003322 | Philippine |
| 3 | Red jungle fowl (G. g. spadiceus) | AP003321 | Laos |
| 4 | Red jungle fowl (G. g. spadiceus) | GU261716 | Myanmar |
| 5 | Red jungle fowl (G. g. spadiceus) | GU261702 | China |
| 6 | Red jungle fowl (G. g. jabouillei) | GU261674 | China |
| 7 | Red jungle fowl (G. g. murghi) | GU261708 | India |
| 8 | Red jungle fowl (G. g. murghi) | KP211423 | India |
| 9 | Green jungle fowl (G. varius) | AP003324 | Indonesia |
| 10 | Grey jungle fowl (G. sonheratii) | AP003320 | Laos |
| 11 | Ceylon jungle fowl (G. lafayettei) | AP003325 | Japan |
| 12 | White Leghorn | AP003317 | Europe |
| 13 | White Plymouth Rock | AP003318 | Europe |
| 14 | Lao’s negative chicken | AP00319 | Laos |
| 15 | Domestic chicken | KY039399 | Philippine |
| 16 | Domestic chicken | KY039425 | Indonesia |
| 17 | Domestic chicken | KY039394 | Long Island New Guinea |
| 18 | Aseel | KP211418 | India |
| 19 | Ghagus | KP211419 | India |
| 20 | Haringhata Black | KP211420 | India |
| 21 | Nicobari Black | KP211421 | India |
| 22 | Nicobari Brown | KP211422 | India |
| 23 | Tellicherry | KP211424 | India |
| 24 | Kadaknath | KP211425 | India |
| 25 | Rugao Yellow | KP742951 | China |
| 26 | Xiaoxiang | KX781319 | China |
| 27 | Xuefeng | GU261675 | China |
| 28 | Lverwu | GU261712 | China |
| 29 | Jinhu Wufeng | KR347464 | China |
| 30 | Tibetan | DQ648776 | China |
| Gene | Strand | Position | Length (bp) | Intergenic Nucleotide * | Codons | Anti-Codon | ||
|---|---|---|---|---|---|---|---|---|
| Start | End | Start | Stop | |||||
| D-loop | H | 1 | 1231 | 1231 | 0 | |||
| tRNA-Phe | H | 1232 | 1301 | 70 | 0 | GAA | ||
| 12S rRNA | H | 1302 | 2276 | 975 | 0 | |||
| tRNA-Val | H | 2277 | 2349 | 73 | 0 | TAC | ||
| 16S rRNA | H | 2350 | 3971 | 1622 | 0 | |||
| tRNA-Leu2 | H | 3972 | 4045 | 74 | 9 | TAA | ||
| ND1 | H | 4055 | 5029 | 975 | 0 | ATG | TAA | |
| tRNA-Ile | H | 5030 | 5101 | 72 | 5 | GAT | ||
| tRNA-Gln | L | 5107 | 5177 | 71 | −1 | TTG | ||
| tRNA-Met | H | 5177 | 5245 | 69 | 0 | CAT | ||
| ND2 | H | 5246 | 6286 | 1041 | −2 | ATG | TAG | |
| tRNA-Trp | H | 6285 | 6360 | 76 | 6 | TCA | ||
| tRNA-Ala | L | 6367 | 6435 | 69 | 3 | TGC | ||
| tRNA-Asn | L | 6439 | 6511 | 73 | 1 | GTT | ||
| tRNA-Cys | L | 6513 | 6578 | 66 | −1 | GCA | ||
| tRNA-Tyr | L | 6578 | 6648 | 71 | 1 | GTA | ||
| COI | H | 6650 | 8200 | 1551 | −9 | GTG | AGG | |
| tRNA-Ser2 | L | 8192 | 8266 | 75 | 2 | TGA | ||
| tRNA-Asp | H | 8269 | 8337 | 69 | 1 | GTC | ||
| COII | H | 8339 | 9022 | 684 | 1 | ATG | TAA | |
| tRNA-Lys | H | 9024 | 9091 | 68 | 1 | TTT | ||
| ATP8 | H | 9093 | 9257 | 165 | −10 | ATG | TAA | |
| ATP6 | H | 9248 | 9931 | 684 | −1 | ATG | TAA | |
| COIII | H | 9931 | 10714 | 784 | 0 | ATG | T-- | |
| tRNA-Gly | H | 10715 | 10783 | 69 | 0 | TCC | ||
| ND3 | H | 10784 | 11135 | 352 | 1 | ATG | TAA | |
| tRNA-Arg | H | 11137 | 11204 | 68 | 0 | TCG | ||
| ND4L | H | 11205 | 11501 | 297 | −7 | ATG | TAA | |
| ND4 | H | 11495 | 12872 | 1378 | 0 | ATG | T-- | |
| tRNA-His | H | 12873 | 12941 | 69 | 1 | GTG | ||
| tRNA-Ser1 | H | 12943 | 13007 | 65 | 1 | GCT | ||
| tRNA-Leu1 | H | 13009 | 13079 | 71 | 0 | TAG | ||
| ND5 | H | 13080 | 14897 | 1818 | 4 | ATG | TAA | |
| Cytb | H | 14902 | 16044 | 1143 | 3 | ATG | TAA | |
| tRNA-Thr | H | 16048 | 16116 | 69 | 0 | TGT | ||
| tRNA-Pro | L | 16117 | 16186 | 70 | 6 | TGG | ||
| ND6 | L | 16193 | 16714 | 522 | 2 | ATG | TAA | |
| tRNA-Glu | L | 16717 | 16784 | 68 | TTC | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, L.D.; Giang, T.T.N.; Nguyen, V.B.; Pham, T.P.M.; Tran, T.T.T.; Nguyen, T.Q.C.; Van Nguyen, K.; Do, D.N. The Complete Mitochondrial Genome and Phylogenetic Analyses of To Chicken in Vietnam. Genes 2023, 14, 1088. https://doi.org/10.3390/genes14051088
Pham LD, Giang TTN, Nguyen VB, Pham TPM, Tran TTT, Nguyen TQC, Van Nguyen K, Do DN. The Complete Mitochondrial Genome and Phylogenetic Analyses of To Chicken in Vietnam. Genes. 2023; 14(5):1088. https://doi.org/10.3390/genes14051088
Chicago/Turabian StylePham, Lan Doan, Thi Thanh Nhan Giang, Van Ba Nguyen, Thi Phuong Mai Pham, Thi Thu Thuy Tran, Thi Quynh Chau Nguyen, Khanh Van Nguyen, and Duy Ngoc Do. 2023. "The Complete Mitochondrial Genome and Phylogenetic Analyses of To Chicken in Vietnam" Genes 14, no. 5: 1088. https://doi.org/10.3390/genes14051088
APA StylePham, L. D., Giang, T. T. N., Nguyen, V. B., Pham, T. P. M., Tran, T. T. T., Nguyen, T. Q. C., Van Nguyen, K., & Do, D. N. (2023). The Complete Mitochondrial Genome and Phylogenetic Analyses of To Chicken in Vietnam. Genes, 14(5), 1088. https://doi.org/10.3390/genes14051088





