The Second Highest Prevalence of Celiac Disease Worldwide: Genetic and Metabolic Insights in Southern Brazilian Mennonites
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Public Involvement
2.2. Epidemiological Study
2.3. Serological Screening
2.4. HLA-DQ2.5/DQ8 Genetic Screening
2.5. Metabolomic Analysis by LC-HRMS
2.6. Statistical and Bioinformatic Analyses
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Ludvigsson, J.F.; Leffler, D.A.; Bai, J.C.; Biagi, F.; Fasano, A.; Green, P.H.R.; Hadjivassiliou, M.; Kaukinen, K.; Kelly, C.P.; Leonard, J.N.; et al. The Oslo definitions for coeliac disease and related terms. Gut 2013, 62, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Bourgey, M.; Calcagno, G.; Tinto, N.; Gennarelli, D.; Margaritte-Jeannin, P.; Greco, L.; Limongelli, M.G.; Esposito, O.; Marano, C.; Troncone, R.; et al. HLA related genetic risk for coeliac disease. Gut 2007, 56, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Rewers, M.; Eisenbarth, G.S. Genetic testing: Who should do the testing and what is the role of genetic testing in the setting of celiac disease? Gastroenterology 2005, 128, S33–S37. [Google Scholar] [CrossRef] [PubMed]
- Kotze, L.M.S. Celiac disease in Brazilian patients: Associations, complications and causes of death. Forty years of clinical experience. Arq. Gastroenterol. 2009, 46, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A.; Catassi, C. Current approaches to diagnosis and treatment of celiac disease: An evolving spectrum. Gastroenterology 2001, 120, 636–651. [Google Scholar] [CrossRef]
- Utiyama, S.R.R.; Nass, F.R.; Kotze, L.M.S.; Nisihara, R.M.; Ambrosio, A.R.; de Messias-Reason, I.T. Triagem sorológica de familiares de pacientes com doença celíaca: Anticorpos anti-endomísio, antitransglutaminase ou ambos? Arq. Gastroenterol. 2007, 44, 156–161. [Google Scholar] [CrossRef]
- Green, P.H.R.; Lebwohl, B.; Greywoode, R. Celiac disease. J. Allergy Clin. Immunol. 2015, 135, 1099–1106. [Google Scholar] [CrossRef]
- Romano, L.; Pellegrino, R.; Sciorio, C.; Barone, B.; Gravina, A.G.; Santonastaso, A.; Mucherino, C.; Astretto, S.; Napolitano, L.; Aveta, A.; et al. Erectile and sexual dysfunction in male and female patients with celiac disease: A cross-sectional observational study. Andrology 2022, 10, 910–918. [Google Scholar] [CrossRef]
- Aaron, L.; Torsten, M.; Patricia, W. Autoimmunity in celiac disease: Extra-intestinal manifestations. Autoimmun. Rev. 2019, 18, 241–246. [Google Scholar] [CrossRef]
- Rodrigo, L.; Beteta-Gorriti, V.; Alvarez, N.; Gómez de Castro, C.; De Dios, A.; Palacios, L.; Santos-Juanes, J. Cutaneous and Mucosal Manifestations Associated with Celiac Disease. Nutrients 2018, 10, 800. [Google Scholar] [CrossRef]
- Lindfors, K.; Ciacci, C.; Kurppa, K.; Lundin, K.E.; Makharia, G.K.; Mearin, M.L.; Murray, J.A.; Verdu, E.F.; Kaukinen, K. Coeliac disease. Nat. Rev. Dis. Prim. 2019, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Ratsch, I.M.; Gandolfi, L.; Pratesi, R.; Fabiani, E.; El Asmar, R.; Frijia, M.; Bearzi, I.; Vizzoni, L. Why is coeliac disease endemic in the people of the Sahara? Lancet 1999, 354, 647–648. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arora, A.; Strand, T.A.; Leffler, D.A.; Catassi, C.; Green, P.H.; Kelly, C.P.; Ahuja, V.; Makharia, G.K. Global Prevalence of Celiac Disease: Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2018, 16, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.A.G. Prevalence of celiac disease in an urban area of Brazil with predominantly European ancestry. WJG 2006, 12, 6546. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.L.; Hou, L.; Boldt, A.B.W.; Kassem, L.; Alves, V.M.; Nardi, A.E.; McMahon, F.J. Finding Rare, Disease-Associated Variants in Isolated Groups: Potential Advantages of Mennonite Populations. Hum. Biol. 2016, 88, 109–120. [Google Scholar] [CrossRef]
- Hou, L.; Kember, R.L.; Roach, J.C.; O’connell, J.R.; Craig, D.W.; Bucan, M.; Scott, W.K.; Pericak-Vance, M.; Haines, J.L.; Crawford, M.H.; et al. A population-specific reference panel empowers genetic studies of Anabaptist populations. Sci. Rep. 2017, 7, 6079. [Google Scholar] [CrossRef]
- Pardo-Seco, J.; Llull, C.; Berardi, G.; Gómez, A.; Andreatta, F.; Martinón-Torres, F.; Toscanini, U.; Salas, A. Genomic continuity of Argentinean Mennonites. Sci. Rep. 2016, 6, 36392. [Google Scholar] [CrossRef]
- Toscanini, U.; Brisighelli, F.; Llull, C.; Berardi, G.; Gómez, A.; Andreatta, F.; Pardo-Seco, J.; Gómez-Carballa, A.; Martinón-Torres, F.; Álvarez-Iglesias, V.; et al. Charting the Y-chromosome ancestry of present-day Argentinean Mennonites. J. Hum. Genet. 2016, 61, 507–513. [Google Scholar] [CrossRef]
- Jaworski, M.A.; Slater, J.D.; Severini, A.; Hennig, K.R.; Mansour, G.; Mehta, J.G.; Jeske, R.; Schlaut, J.; Pak, C.Y.; Yoon, J.W. Unusual clustering of diseases in a Canadian Old Colony (Chortitza) Mennonite kindred and community. CMAJ 1988, 138, 1017–1025. [Google Scholar]
- Boschmann, S.E.; Boldt, A.B.; de Souza, I.R.; Petzl-Erler, M.L.; Messias-Reason, I.J. The Frequency of the LCT*-13910C>T Polymorphism Associated with Lactase Persistence Diverges among Euro-Descendant Groups from Brazil. Med. Princ. Pract. 2016, 25, 18–20. [Google Scholar] [CrossRef]
- Tursi, A.; Elisei, W.; Giorgetti, G.M.; Gaspardone, A.; Lecca, P.G.; Di Cesare, L.; Brandimarte, G. Prevalence of celiac disease and symptoms in relatives of patients with celiac disease. Eur. Rev. Med. Pharm. Sci. 2010, 14, 567–572. [Google Scholar]
- Al-Toma, A.; Volta, U.; Auricchio, R.; Castillejo, G.; Sanders, D.S.; Cellier, C.; Mulder, C.J.; Lundin, K.E.A. European Society for the Study of Coeliac Disease (ESsCD) guideline for coeliac disease and other gluten-related disorders. United Eur. Gastroenterol. J. 2019, 7, 583–613. [Google Scholar] [CrossRef] [PubMed]
- Volta, U.; Molinaro, N.; De Franceschi, L.; Fratangelo, D.; Bianchi, F.B. IgA anti-endomysial antibodies on human umbilical cord tissue for celiac disease screening. Dig. Dis. Sci. 1995, 40, 1902–1905. [Google Scholar] [CrossRef] [PubMed]
- Marsh, M.N. Gluten, major histocompatibility complex, and the small intestine. Gastroenterology 1992, 102, 330–354. [Google Scholar] [CrossRef] [PubMed]
- Olerup, O.; Aldener, A.; Fogdell, A. HLA-DQB1 and -DQA1 typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Tissue Antigens 1993, 41, 119–134. [Google Scholar] [CrossRef]
- Profaizer, T.; Eckels, D.; Delgado, J.C. Celiac disease and HLA typing using real-time PCR with melting curve analysis. Tissue Antigens 2011, 78, 31–37. [Google Scholar] [CrossRef]
- Vorkas, P.A.; Isaac, G.; Anwar, M.A.; Davies, A.H.; Want, E.J.; Nicholson, J.K.; Holmes, E. Untargeted UPLC-MS Profiling Pipeline to Expand Tissue Metabolome Coverage: Application to Cardiovascular Disease. Anal. Chem. 2015, 87, 4184–4193. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- González-Galarza, F.F.; Takeshita, L.Y.; Santos, E.J.; Kempson, F.; Maia, M.H.T.; Da Silva, A.L.S.; Silva, A.L.T.E.; Ghattaoraya, G.; Alfirevic, A.; Jones, A.; et al. Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015, 43, D784–D788. [Google Scholar] [CrossRef]
- Fairweather, D.; Frisancho-Kiss, S.; Rose, N.R. Sex Differences in Autoimmune Disease from a Pathological Perspective. Am. J. Pathol. 2008, 173, 600–609. [Google Scholar] [CrossRef]
- Csizmadia, C.G.; Mearin, M.L.; von Blomberg, B.M.; Brand, R.; Verloove-Vanhorick, S.P. An iceberg of childhood coeliac disease in the Netherlands. Lancet 1999, 353, 813–814. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, S.; Alliet, P.; Gillis, P.; Declercq, P.; Mewis, A. Seroprevalence of celiac disease in Belgian children and adolescents. Acta Gastroenterol. Belg. 2012, 75, 325–330. [Google Scholar] [PubMed]
- Händel, N.; Mothes, T.; Petroff, D.; Baber, R.; Jurkutat, A.; Flemming, G.; Kiess, W.; Hiemisch, A.; Körner, A.; Schlumberger, W.; et al. Will the Real Coeliac Disease Please Stand Up? Coeliac Disease Prevalence in the German LIFE Child Study. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Gujral, N. Celiac disease: Prevalence, diagnosis, pathogenesis and treatment. World J. Gastroenterol. WJG 2012, 18, 6036. [Google Scholar] [CrossRef] [PubMed]
- Alencar, M.L.; Ortiz-Agostinho, C.L.; Damião, A.O.; Abrantes-Lemos, C.P.; Leite, A.Z.; Brito, T.D.; Chamone, D.D.; Silva, M.E.; Giannella-Neto, D.; Sipahi, A.M. Prevalence of celiac disease among blood donors in SÃO PAULO—The most populated city in Brazil. Clinics 2012, 67, 1013–1018. [Google Scholar] [CrossRef]
- Kratzer, W. Prevalence of celiac disease in Germany: A prospective follow-up study. World J. Gastroenterol. WJG 2013, 19, 2612. [Google Scholar] [CrossRef]
- Vilppula, A.; Collin, P.; Mäki, M.; Valve, R.; Luostarinen, M.; Krekelä, I.; Patrikainen, H.; Kaukinen, K. Undetected coeliac disease in the elderly. Dig. Liver Dis. 2008, 40, 809–813. [Google Scholar] [CrossRef]
- Mäki, M.; Mustalahti, K.; Kokkonen, J.; Kulmala, P.; Haapalahti, M.; Karttunen, T.; Ilonen, J.; Laurila, K.; Dahlbom, I.; Hansson, T.; et al. Prevalence of Celiac Disease among Children in Finland. N. Engl. J. Med. 2003, 348, 2517–2524. [Google Scholar] [CrossRef]
- Gomez, J.C.; Selvaggio, G.S.; Viola, M.; Pizarro, B.; Motta, G.; Barrio, S.; Castelletto, R.; Echeverria, R.; Sugai, E.; Vazquez, H.; et al. Prevalence of celiac disease in argentina: Screening of an adult population in the La Plata area. Am. J. Gastroenterol. 2001, 96, 2700–2704. [Google Scholar] [CrossRef]
- Cárdenas-Roldán, J.; Rojas-Villarraga, A.; Anaya, J.M. How do autoimmune diseases cluster in families? A systematic review and meta-analysis. BMC Med. 2013, 11, 73. [Google Scholar] [CrossRef]
- Parra-Medina, R.; Molano-Gonzalez, N.; Rojas-Villarraga, A.; Agmon-Levin, N.; Arango, M.-T.; Shoenfeld, Y.; Anaya, J.-M. Prevalence of Celiac Disease in Latin America: A Systematic Review and Meta-Regression. PLoS ONE 2015, 10, e0124040. [Google Scholar] [CrossRef] [PubMed]
- Mearin, M.L. The prevention of coeliac disease. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 493–501. [Google Scholar] [CrossRef]
- Högberg, L.; Fälth-Magnusson, K.; Grodzinsky, E.; Stenhammar, L. Familial Prevalence of Coeliac Disease: A Twenty-Year Follow-up Study. Scand. J. Gastroenterol. 2003, 38, 61–65. [Google Scholar] [CrossRef]
- Ciclitira, P.J.; Johnson, M.W.; Dewar, D.H.; Ellis, H.J. The pathogenesis of coeliac disease. Mol. Asp. Med. 2005, 26, 421–458. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arora, S.; Lal, S.; Strand, T.A.; Makharia, G.K. Risk of Celiac Disease in the First- and Second-Degree Relatives of Patients with Celiac Disease: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2015, 110, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Karell, K.; Louka, A.S.; Moodie, S.J.; Ascher, H.; Clot, F.; Greco, L.; Ciclitira, P.J.; Sollid, L.M.; Partanen, J. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: Results from the European genetics cluster on celiac disease. Hum. Immunol. 2003, 64, 469–477. [Google Scholar] [CrossRef]
- Basturk, A.; Artan, R.; Yilmaz, A. The incidence of HLA-DQ2/DQ8 in Turkish children with celiac disease and a comparison of the geographical distribution of HLA-DQ. Gastroenterol. Rev. 2017, 4, 256–261. [Google Scholar] [CrossRef]
- Kotze, L.M.S.; Nisihara, R.; Utiyama, S.R.R.; Kotze, L.R. Absence of HLA-DQ2 and HLA-DQ8 does not exclude celiac disease in Brazilian patients. Rev. Esp. Enferm. Dig. 2014, 106, 561–562. [Google Scholar]
- Ricaño-Ponce, I.; Wijmenga, C.; Gutierrez-Achury, J. Genetics of celiac disease. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 399–412. [Google Scholar] [CrossRef]
- Trynka, G.; Hunt, K.A.; Bockett, N.A.; Romanos, J.; Mistry, V.; Szperl, A.; Bakker, S.F.; Bardella, M.T.; Bhaw-Rosun, L.; Castillejo, G.; et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011, 43, 1193–1201. [Google Scholar] [CrossRef]
- Hua, L.; Xiang, S.; Xu, R.; Xu, X.; Liu, T.; Shi, Y.; Wu, L.; Wang, R.; Sun, Q. Causal association between rheumatoid arthritis and celiac disease: A bidirectional two-sample mendelian randomization study. Front. Genet. 2022, 13, 976579. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Cui, C.; Li, N.; Sun, X.; Wen, L.; Gao, E.; Wang, F. Antioxidant activity and protective effect of wheat germ peptides in an in vitro celiac disease model via Keap1/Nrf2 signaling pathway. Food Res. Int. 2022, 161, 111864. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione Metabolism and Its Implications for Health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Wahab, P.J.; Peters, W.H.M.; Roelofs, H.M.J.; Jansen, J.B.M.J. Glutathione S-Transferases in Small Intestinal Mucosa of Patients with Coeliac Disease. Jpn. J. Cancer Res. 2001, 92, 279–284. [Google Scholar] [CrossRef]
- Coles, B.F.; Chen, G.; Kadlubar Fred, F.; Radominska-Pandya, A. Interindividual variation and organ-specific patterns of glutathione S-transferase alpha, mu, and pi expression in gastrointestinal tract mucosa of normal individuals. Arch. Biochem. Biophys. 2002, 403, 270–276. [Google Scholar] [CrossRef]
- Sevinc, E.; Sevinc, N.; Akar, H.H.; Ozelcoskun, B.D.; Sezgin, G.C.; Arslan, D.; Kendirci, M. Plasma glutamine and cystine are decreased and negatively correlated with endomysial antibody in children with celiac disease. Asia Pac. J. Clin. Nutr. 2016, 25, 452–456. [Google Scholar]
- Moretti, S.; Mrakic-Sposta, S.; Roncoroni, L.; Vezzoli, A.; Dellanoce, C.; Monguzzi, E.; Branchi, F.; Ferretti, F.; Lombardo, V.; Doneda, L.; et al. Oxidative stress as a biomarker for monitoring treated celiac disease. Clin. Transl. Gastroenterol. 2018, 9, e157. [Google Scholar] [CrossRef]
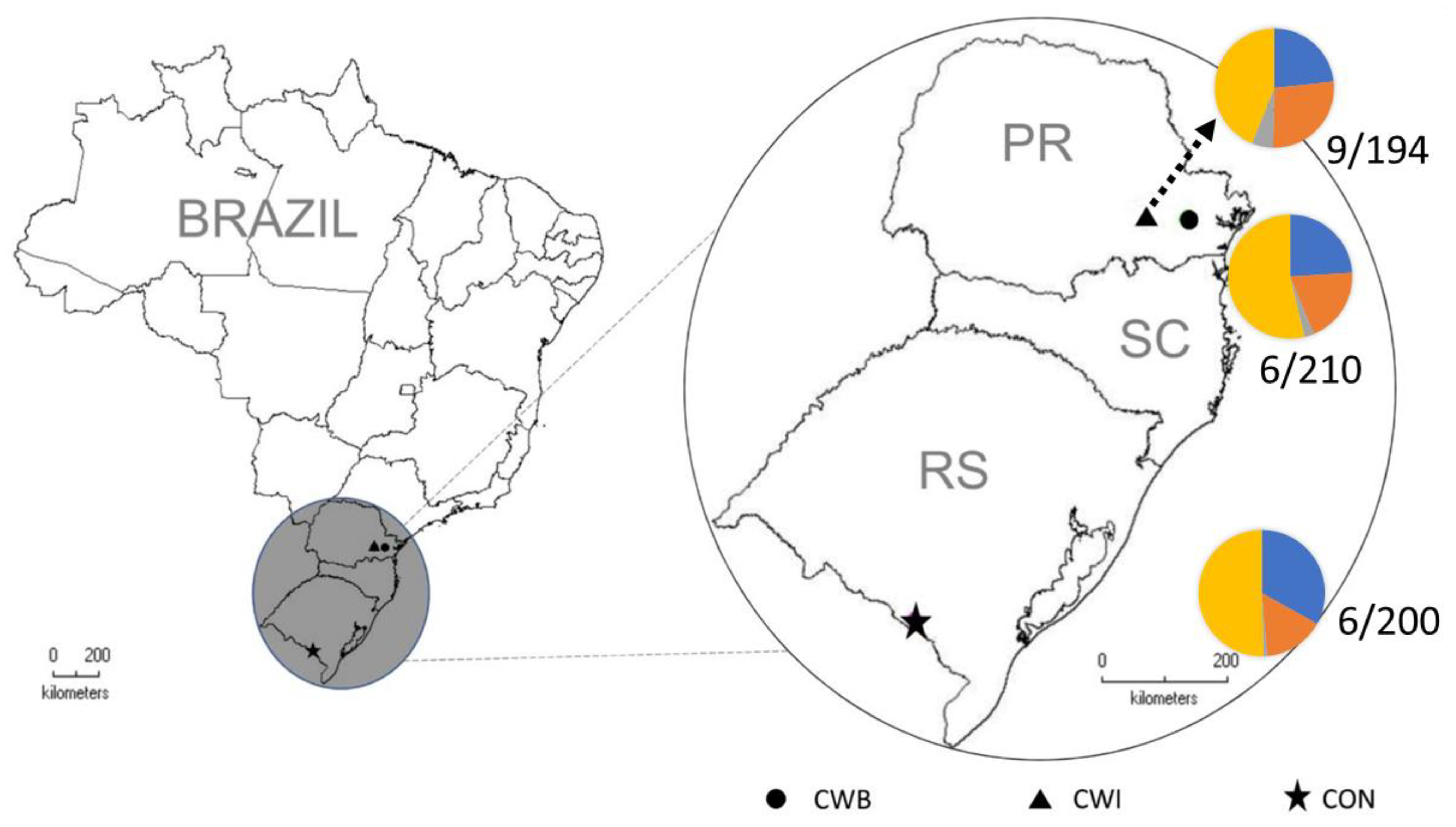
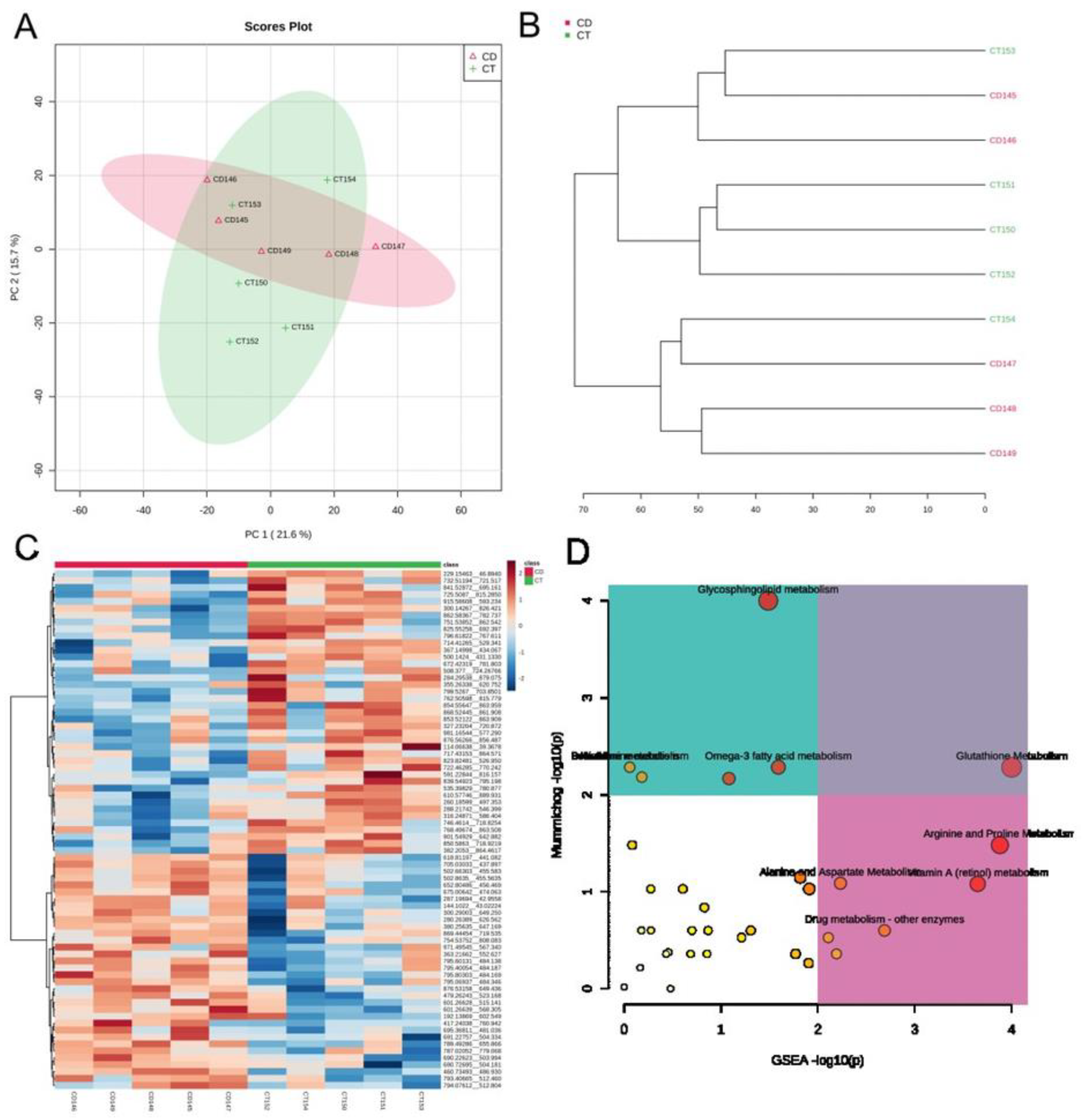
| CON | CWI | CTB | p Value | Total | CD Patients | Non-CD Patients | p Value | |
|---|---|---|---|---|---|---|---|---|
| N | N = 200 | N = 210 | N = 194 | N = 604 | N = 21 | N = 583 | ||
| Gender (F/M) | 120/80 | 136/74 | 107/87 | 0.143 | 363/241 | 16/05 | 347/236 | 0.173 |
| Median (min-max) | Median (min-max) | Median (min-max) | Median (min-max) | Median (min-max) | Median (min-max) | |||
| Age–female (years) | 57.0 (14.0–83.0) | 46.0 (14.4–89.0) | 44.2 (14.0–89.2) | 0.003 q = 0.026 | 49.0 (14.0–89.2) | 32.0 (17.1–69.0) | 50.0 (14.0–89.2) | 0.015 q = 0.039 |
| Age–male (years) | 54.0 (17.0–82.0) | 53.7 (12.0–92.1) | 49.5 (15.9–88.0) | 0.241 | 51.0 (12.0–92.1) | 64.6 (54.9–75.0) | 51.0 (12.0–92.1) | 0.095 |
| BMI–female | 27.6 (15.8–45.8) | 24.6 (16.2–35.2) | 23.6 (15.1–40.6) | 1 × 10−4 q = 0.013 | 25.2 (15.1–45.8) | 22.4 (19.6–45.8) | 25.5 (15.1–44.5) | 0.235 |
| BMI–male | 28.4 (19.2–42.9) | 26.2 (16.1–47.6) | 26.3 (16.6–38.7) | 4 × 10−4 q = 0.015 | 27.1 (16.1–47.6) | 27.4 (26.0–28.9) | 26.9 (16.1–47.6) | 0.724 |
| % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | |||
| Symptoms/diagnoses | ||||||||
| Chronic diarrhea | 2.11 (4/190) | 7.14 (13/182) | 5.70 (9/158) | 0.068 | 4.90 (26/530) | 19.05 (4/21) | 4.32 (22/509) | 0.015 q = 0.010 |
| Weight loss | 3.68 (7/190) | 7.94 (10/126) | 5.06 (8/158) | 0.251 | 5.27 (25/474) | 15.00 (3/20) | 4.85 (22/454) | 0.081 |
| Iron-deficiency anemia | 7.37 (14/190) | 10.94 (14/128) | 8.23 (13/158) | 0.527 | 8.61 (41/476) | 38.09 (8/21) | 7.25 (33/455) | 1 × 10−4 q = 0.018 |
| Bloating | 31.58 (60/190) | 28.96 (53/183) | 27.21 (43/158) | 0.663 | 29.37 (156/531) | 38.09 (8/21) | 29.02 (148/510) | 0.462 |
| Constipation | 12.63 (24/190) | 19.04 (24/126) | 20.88 (33/158) | 0.099 | 17.09 (81/474) | 10.00 (2/20) | 17.40 (79/454) | 0.549 |
| Chronic abdominal pain | 16.32 (31/190) | 16.48 (30/182) | 15.28 (24/157) | 0.951 | 16.07 (85/529) | 42.85 (9/21) | 14.96 (76/508) | 0.002 q = 0.021 |
| Joint pain | 26.84 (51/190) | 9.52 (12/126) | 8.33 (13/156) | 1 × 10−6 q = 0.002 | 16.10 (76/472) | 5.00 (1/20) | 16.59 (75/452) | 0.223 |
| Diabetes mellitus type 1 | 1.06 (2/188) | 0.79 (1/126) | 2.46 (3/122) | n.c. | 1.38 (6/436) | 0 (0/19) | 1.44 (6/417) | nc |
| Irritable bowel syndrome | 5.82 (11/189) | 5.00 (6/120) | 4.04 (4/99) | 0.887 | 4.90 (20/408) | 6.25 (1/16) | 5.10 (20/392) | 1 |
| Lactose intolerance | 1.62 (3/185) | 3.35 (6/179) | 9.59 (14/146) | 0.003 q = 0.028 | 4.51 (23/510) | 4.76 (1/21) | 4.50 (22/489) | 1 |
| Autoimmune thyroiditis | 10.47 (20/191) | 11.29 (14/124) | 14.74 (23/156) | 0.454 | 12.10 (57/471) | 15.00 (3/20) | 12.36 (56/453) | 0.727 |
| Rheumatoid arthritis | 11.30 (20/177) | 4.00 (5/125) | 5.52 (9/163) | 0.031 q = 0.047 | 7.31 (34/465) | 0 (0/19) | 7.62 (34/446) | nc |
| Celiac disease | 3.00 (6/200) | 2.86 (6/210) | 4.64 (9/194) | 0.560 | 3.48 (21/604) | - | - | |
| Any autoimmune disease * | 20.90 (37/177) | 15.08 (19/126) | 20.24 (33/163) | 0.400 | 19.10 (89/466) | 15.79 (3/19) * | 19.24 (86/447) * | 1 |
| Familiar aggregation | ||||||||
| FDR with CD | 3.21 (6/187) | 11.21 (12/107) | 5.03 (8/159) | 0.015 q = 0.044 | 5.74 (26/453) | 31.58 (6/19) | 4.61 (20/434) | 3 × 10−4 q = 0.015 |
| SDR with CD | 1.60 (3/187) | 4.81 (5/104) | 4.43 (7/158) | 0.221 | 3.34 (15/449) | 21.05 (4/19) | 2.56 (11/430) | 0.002 q = 0.023 |
| Any relative with CD | 5.88 (11/187) | 14.95 (16/107) | 6.29 (10/159) | 0.013 q = 0.036 | 8.17 (37/453) | 36.84 (7/19) | 4.61 (20/434) | 3 × 10−5 q = 0.010 |
| HLA | N = 115 | N = 155 | N = 121 | N = 391 | N = 13 | N = 378 | ||
| DQ2.5 (+) | 33.91 (39) | 29.03 (45) | 26.45 (32) | 0.559 | 29.67 (116) | 92.31 (12) | 27.51 (104) | 3 × 10−6 q = 0.007 |
| Only DQ2.5 (+) | 33.04 (38) | 23.23 (36) | 23.97 (29) | 0.159 | 26.09 (102) | 92.31 (12) | 24.07 (90) | 1 × 10−6 q = 0.005 |
| DQ8 (+) | 16.52 (19) | 32.90 (51) | 22.31 (27) | 0.007 q = 0.034 | 24.81 (97) | 0 | 25.66 (97) | 0.044 q = 0.05 |
| Only DQ8 (+) | 15.65 (18) | 27.10 (42) | 19.83 (24) | 0.072 | 21.48 (84) | 0 | 22.22 (84) | 0.080 |
| DQ2.5 (+) and/or DQ8 (+) | 49.57 (57) | 56.13 (87) | 46.28 (56) | 0.248 | 51.15 (200) | 92.31 (12) | 49.74 (188) | 0.003 q = 0.031 |
| Symptoms | CD% (n/N) | Non-CD% (n/N) | OR | 95% CI | p Value |
|---|---|---|---|---|---|
| Chronic diarrhea | 19.05 (4/21) | 4.32 (22/509) | 5.20 | 1.61–16.78 | 0.006 |
| Iron-deficiency anemia | 38.09 (8/21) | 7.25 (33/455) | 7.86 | 3.04–20.33 | <10−3 |
| Chronic abdominal pain | 42.85 (9/21) | 14.96 (76/508) | 4.26 | 1.73–10.46 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, L.C.; Dornelles, A.C.; Nisihara, R.M.; Bruginski, E.R.D.; Santos, P.I.d.; Cipolla, G.A.; Boschmann, S.E.; Messias-Reason, I.J.d.; Campos, F.R.; Petzl-Erler, M.L.; et al. The Second Highest Prevalence of Celiac Disease Worldwide: Genetic and Metabolic Insights in Southern Brazilian Mennonites. Genes 2023, 14, 1026. https://doi.org/10.3390/genes14051026
Oliveira LC, Dornelles AC, Nisihara RM, Bruginski ERD, Santos PId, Cipolla GA, Boschmann SE, Messias-Reason IJd, Campos FR, Petzl-Erler ML, et al. The Second Highest Prevalence of Celiac Disease Worldwide: Genetic and Metabolic Insights in Southern Brazilian Mennonites. Genes. 2023; 14(5):1026. https://doi.org/10.3390/genes14051026
Chicago/Turabian StyleOliveira, Luana Caroline, Amanda Coelho Dornelles, Renato Mitsunori Nisihara, Estevan Rafael Dutra Bruginski, Priscila Ianzen dos Santos, Gabriel Adelman Cipolla, Stefanie Epp Boschmann, Iara José de Messias-Reason, Francinete Ramos Campos, Maria Luiza Petzl-Erler, and et al. 2023. "The Second Highest Prevalence of Celiac Disease Worldwide: Genetic and Metabolic Insights in Southern Brazilian Mennonites" Genes 14, no. 5: 1026. https://doi.org/10.3390/genes14051026
APA StyleOliveira, L. C., Dornelles, A. C., Nisihara, R. M., Bruginski, E. R. D., Santos, P. I. d., Cipolla, G. A., Boschmann, S. E., Messias-Reason, I. J. d., Campos, F. R., Petzl-Erler, M. L., & Boldt, A. B. W. (2023). The Second Highest Prevalence of Celiac Disease Worldwide: Genetic and Metabolic Insights in Southern Brazilian Mennonites. Genes, 14(5), 1026. https://doi.org/10.3390/genes14051026






