Organization and Characterization of the Promoter Elements of the rRNA Operons in the Slow-Growing Pathogen Mycobacterium kumamotonense
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. DNA Isolation
2.3. Cloning and Sequencing of rRNA Operons of M. kumamotonense
2.4. Cell Line Culture and Infection
2.5. RNA Isolation and Synthesis of cDNA
2.6. Rapid Amplification of cDNA Ends (RACE)
2.7. Analysis of Precursor rRNA (pre-rrn) by RT-qPCR
2.8. Statistical Analysis
2.9. Alignment of Sequences
3. Results
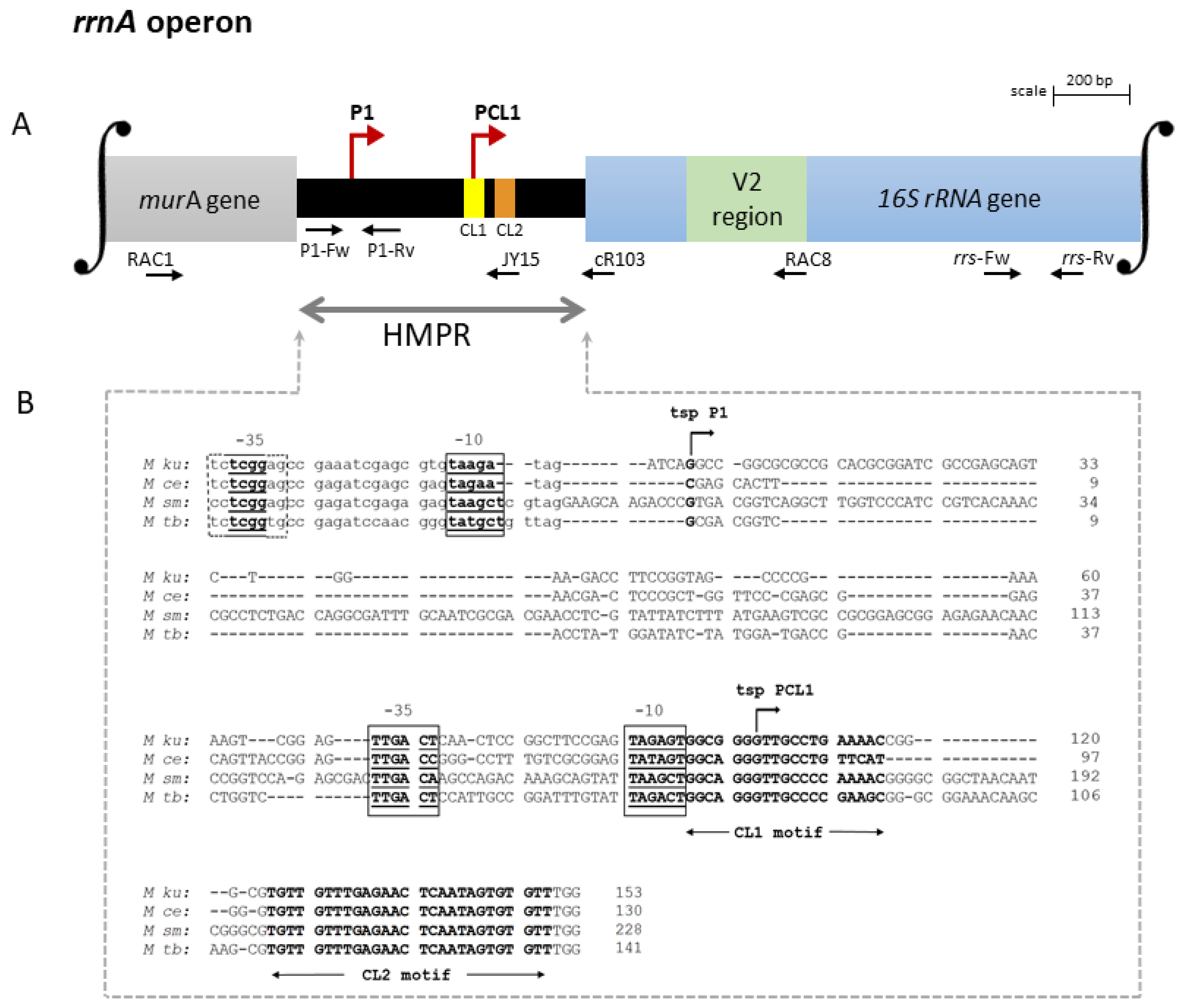
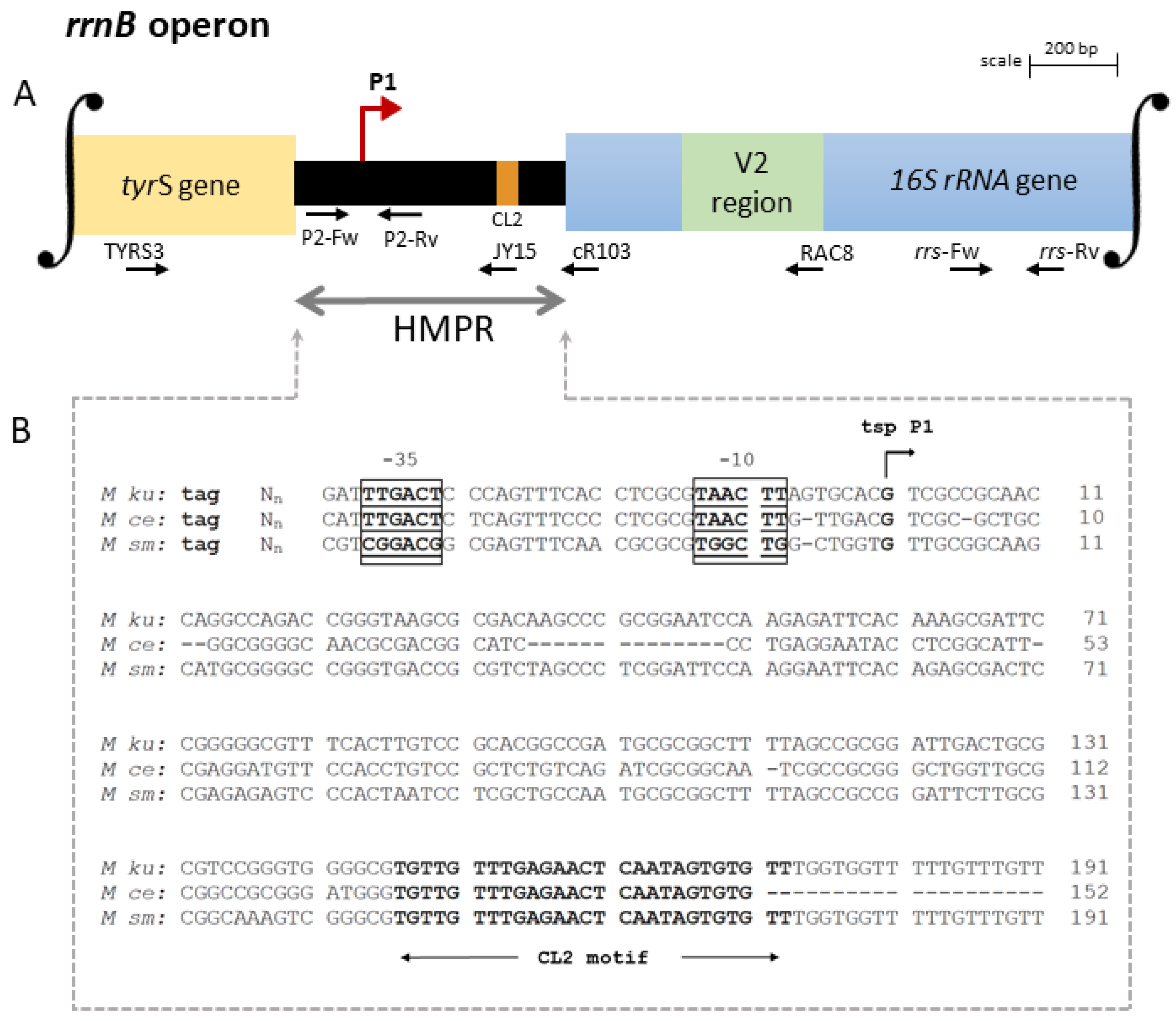
3.1. Organization of the rRNA Promoters
3.2. Expression of the Genes Related to Dormancy
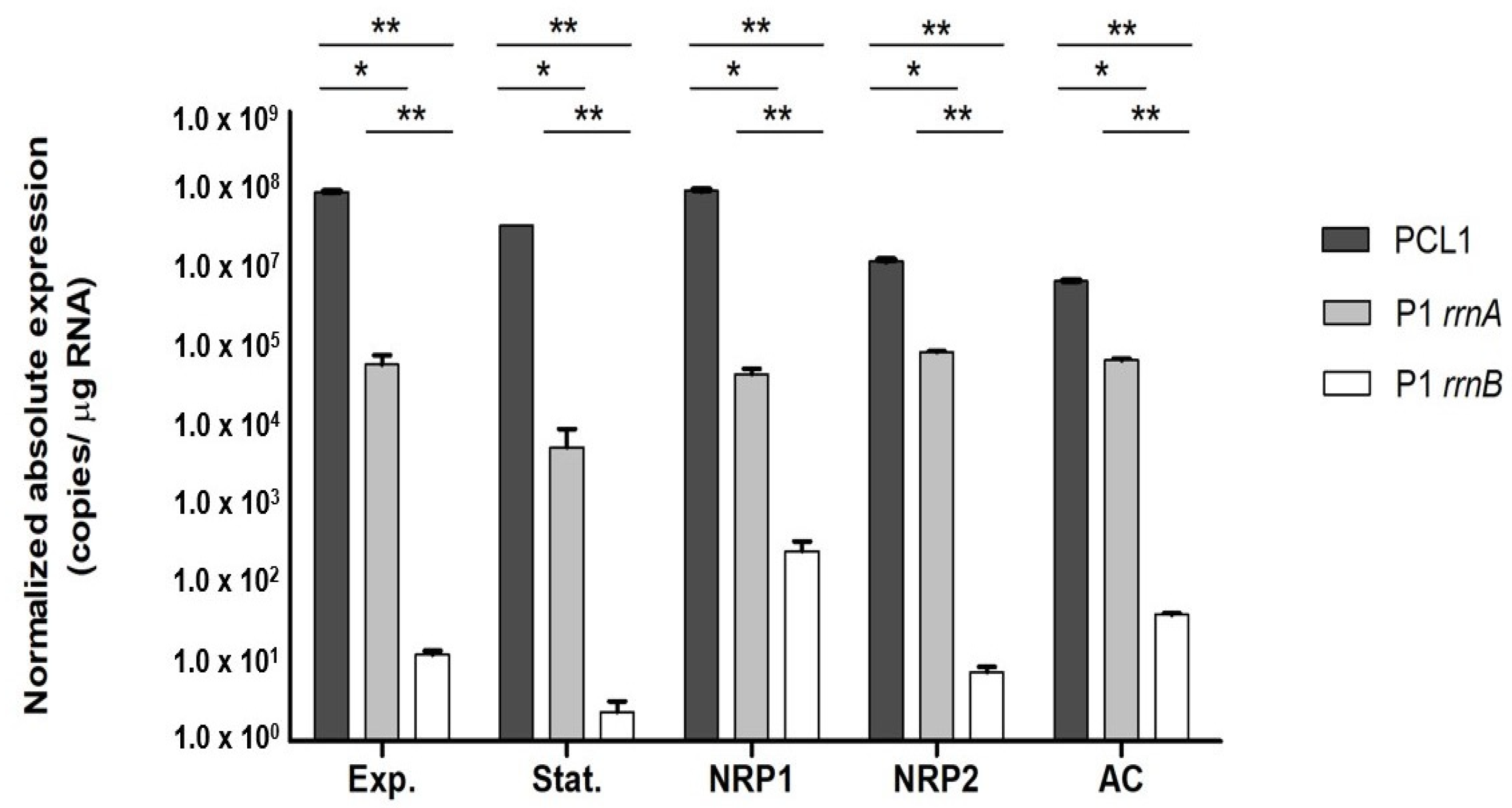
3.3. Expression of the Pre-rrn Products under Stress Conditions
4. Discussion
4.1. Arrangement of the rrnA and rrnB Promoters
4.2. Contribution of the rrn Promoters to rRNA Synthesis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Masaki, T.; Ohkusu, K.; Hata, H.; Fujiwara, N.; Iihara, H.; Yamada-Noda, M.; Nhung, P.H.; Hayashi, M.; Asano, Y.; Kawamura, Y.; et al. Mycobacterium kumamotonense Sp. Nov. Recovered from Clinical Specimen and the First Isolation Report of Mycobacterium arupense in Japan: Novel Slowly Growing, Nonchromogenic Clinical Isolates Related to Mycobacterium terrae Complex. Microbiol. Immunol. 2006, 50, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Aranda, A.; Jimenez, M.S.; Yubero, J.; Chaves, F.; Rubio-Garcia, R.; Palenque, E.; Garcia, M.J.; Menendez, M.C. Misindentification of Mycobacterium kumamotonense as M. tuberculosis. Emerg. Infect. Dis. 2010, 16, 1178–1180. [Google Scholar] [CrossRef]
- Kontos, F.; Mavromanolakis, D.N.; Zande, M.C.; Gitti, Z.G. Isolation of Mycobacterium kumamotonense from a Patient with Pulmonary Infection and Latent Tuberculosis. Indian J. Med. Microbiol. 2016, 34, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Iemura-Kashiwagi, M.; Ito, I.; Ikeguchi, R.; Kadoya, M.; Iemura, T.; Yoshida, S.; Suzuki, K.; Hirai, T. Soft Tissue Infection Caused by Mycolicibacter kumamotonensis. J. Infect. Chemother. 2020, 26, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Solis, A.H.; Gonzalez-Villa, M.; Ramirez-Gonzalez, J.E.; Colin-Munoz, Y.; Cicero-Sabido, R. Mycobacterium kumamotonense in the Cervical Region in an Immunocompetent Patient, Clinical Case Report in Mexico. J. Infect. Dev. Ctries. 2019, 13, 1165–1169. [Google Scholar] [CrossRef]
- Manika, K.; Kontos, F.; Papavasileiou, A.; Papaventsis, D.; Sionidou, M.; Kioumis, I. Severe Pulmonary Disease Caused by Mycolicibacter kumamotonensis. Emerg. Infect. Dis. 2021, 27, 962–964. [Google Scholar] [CrossRef]
- Davarpanah, M.; Azadi, D.; Shojaei, H. Prevalence and Molecular Characterization of Non-tuberculous Mycobacteria in Hospital Soil and Dust of a Developing Country, Iran. Microbiology 2019, 165, 1306–1314. [Google Scholar] [CrossRef]
- Devulder, G.; de Montclos, M.P.; Flandrois, J.P. A Multigene Approach to Phylogenetic Analysis Using the Genus Mycobacterium as a Model. Int. J. Syst. Evol. Microbiol. 2005, 55, 293–302. [Google Scholar] [CrossRef]
- Gonzalez-y-Merchand, J.A.; Colston, M.J.; Cox, R.A. Effects of Growth Conditions on Expression of Mycobacterial murA and tyrS Genes and Contributions of their Transcripts to Precursor rRNA Synthesis. J. Bacteriol. 1999, 181, 4617–4627. [Google Scholar] [CrossRef]
- Gonzalez-y-Merchand, J.A.; Garcia, M.J.; Gonzalez-Rico, S.; Colston, M.J.; Cox, R.A. Strategies Used by Pathogenic and Nonpathogenic Mycobacteria to Synthesize rRNA. J. Bacteriol. 1997, 179, 6949–6958. [Google Scholar] [CrossRef] [PubMed]
- Menendez, M.C.; Garcia, M.J.; Navarro, M.C.; Gonzalez-y-Merchand, J.A.; Rivera-Gutierrez, S.; Garcia-Sanchez, L.; Cox, R.A. Characterization of an rRNA operon (rrnB) of Mycobacterium fortuitum and other mycobacterial species: Implications for the classification of mycobacteria. J. Bacteriol. 2002, 184, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Menendez, M.C.; Yubero, J.; García, M.J.; Jimenez, M.S. Mycobacterium kumamotonense, another member of the Mycobacterium terrae complex unusually carrying two copies of the ribosomal RNA operon. Mycobac. Dis. 2014, 4, 176. [Google Scholar] [CrossRef]
- Wayne, L.G.; Hayes, L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996, 64, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Badillo-Lopez, C.; Gonzalez-Mejia, A.; Helguera-Repetto, A.C.; Salas-Rangel, L.P.; Rivera-Gutierrez, S.; Cerna-Cortes, J.F.; Gonzalez, Y.M.J.A. Differential expression of dnaA and dosR genes among members of the Mycobacterium tuberculosis complex under oxic and hypoxic conditions. Int. Microbiol. 2010, 13, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Soto-Ramirez, M.D.; Aguilar-Ayala, D.A.; Garcia-Morales, L.; Rodriguez-Peredo, S.M.; Badillo-Lopez, C.; Rios-Muniz, D.E.; Meza-Segura, M.A.; Rivera-Morales, G.Y.; Leon-Solis, L.; Cerna-Cortes, J.F.; et al. Cholesterol plays a larger role during Mycobacterium tuberculosis in vitro dormancy and reactivation than previously suspected. Tuberculosis 2017, 103, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chubukov, V.; Sauer, U. Environmental dependence of stationary-phase metabolism in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 2901–2909. [Google Scholar] [CrossRef]
- Miles, A.A.; Misra, S.S.; Irwin, J.O. The estimation of the bactericidal power of the blood. J. Hyg. 1938, 38, 732–749. [Google Scholar] [CrossRef]
- van Soolingen, D.; de Haas, P.E.; Hermans, P.W.; van Embden, J.D. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1994, 235, 196–205. [Google Scholar] [CrossRef]
- Alteri, C.J.; Rios-Sarabia, N.; De la Cruz, M.A.; Gonzalez, Y.M.J.A.; Soria-Bustos, J.; Maldonado-Bernal, C.; Cedillo, M.L.; Yanez-Santos, J.A.; Martinez-Laguna, Y.; Torres, J.; et al. The Flp type IV pilus operon of Mycobacterium tuberculosis is expressed upon interaction with macrophages and alveolar epithelial cells. Front. Cell. Infect. Microbiol. 2022, 12, 916247. [Google Scholar] [CrossRef] [PubMed]
- Jahn, C.E.; Charkowski, A.O.; Willis, D.K. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J. Microbiol. Methods 2008, 75, 318–324. [Google Scholar] [CrossRef]
- Aranda, P.S.; LaJoie, D.M.; Jorcyk, C.L. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis 2012, 33, 366–369. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Stadthagen-Gomez, G.; Helguera-Repetto, A.C.; Cerna-Cortes, J.F.; Goldstein, R.A.; Cox, R.A.; Gonzalez-y-Merchand, J.A. The organization of two rRNA (rrn) operons of the slow-growing pathogen Mycobacterium celatum provides key insights into mycobacterial evolution. FEMS Microbiol. Lett. 2008, 280, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Karboul, A.; Gey van Pittius, N.C.; Namouchi, A.; Vincent, V.; Sola, C.; Rastogi, N.; Suffys, P.; Fabre, M.; Cataldi, A.; Huard, R.C.; et al. Insights into the evolutionary history of tubercle bacilli as disclosed by genetic rearrangements within a PE_PGRS duplicated gene pair. BMC Evol. Biol. 2006, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, Y.M.J.A.; Colston, M.J.; Cox, R.A. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: Comparison of promoter elements and of neighbouring upstream genes. Microbiology 1996, 142 Pt 3, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Menendez Mdel, C.; Rebollo, M.J.; Nunez Mdel, C.; Cox, R.A.; Garcia, M.J. Analysis of the precursor rRNA fractions of rapidly growing mycobacteria: Quantification by methods that include the use of a promoter (rRNA P1) as a novel standard. J. Bacteriol. 2005, 187, 534–543. [Google Scholar] [CrossRef]
- Timm, J.; Gomez, M.; Smith, I. Gene expression and regulation. In Mycobacteria: Molecular Biology and Virulence; Ratledge, J., Dale, W., Eds.; Blackwell Science Ltd.: London, UK, 1999; pp. 59–92. [Google Scholar]
- Manganelli, R. Sigma Factors: Key Molecules in Mycobacterium tuberculosis Physiology and Virulence. Microbiol. Spectr. 2014, 2, 135–160. [Google Scholar] [CrossRef]
- Newton-Foot, M.; Gey van Pittius, N.C. The complex architecture of mycobacterial promoters. Tuberculosis 2013, 93, 60–74. [Google Scholar] [CrossRef]
- Ares, M.A.; Rios-Sarabia, N.; De la Cruz, M.A.; Rivera-Gutierrez, S.; Garcia-Morales, L.; Leon-Solis, L.; Espitia, C.; Pacheco, S.; Cerna-Cortes, J.F.; Helguera-Repetto, C.A.; et al. The sigma factor SigD of Mycobacterium tuberculosis putatively enhances gene expression of the septum site determining protein under stressful environments. New Microbiol. 2017, 40, 199–204. [Google Scholar] [PubMed]
- Condon, C.; Squires, C.; Squires, C.L. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 1995, 59, 623–645. [Google Scholar] [CrossRef]
- Trauner, A.; Lougheed, K.E.; Bennett, M.H.; Hingley-Wilson, S.M.; Williams, H.D. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J. Biol. Chem. 2012, 287, 24053–24063. [Google Scholar] [CrossRef]
- Sherman, D.R.; Voskuil, M.; Schnappinger, D.; Liao, R.; Harrell, M.I.; Schoolnik, G.K. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding Alpha-crystallin. Proc. Natl. Acad. Sci. USA 2001, 98, 7534–7539, Erratum in Proc. Natl. Acad. Sci. USA 2001, 98, 15393. [Google Scholar] [CrossRef] [PubMed]
- Del Portillo, P.; García-Morales, L.; Menéndez, M.C.; Anzola, J.M.; Rodríguez, J.G.; Helguera-Repetto, A.C.; Ares, M.A.; Prados-Rosales, R.; Gonzalez-Y-Merchand, J.A.; García, M.J. Hypoxia Is Not a Main Stress When Mycobacterium tuberculosis Is in a Dormancy-Like Long-Chain Fatty Acid Environment. Front. Cell. Infect. Microbiol. 2019, 8, 449. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.C.; Lukey, P.T.; Robb, L.C.; McAdam, R.A.; Duncan, K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002, 43, 717–731. [Google Scholar] [CrossRef]
- Muttucumaru, D.G.; Roberts, G.; Hinds, J.; Stabler, R.A.; Parish, T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis 2004, 84, 239–246. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Estrada, R.; Méndez-Guerrero, O.; García-Morales, L.; González-y-Merchand, J.A.; Cerna-Cortes, J.F.; Menendez, M.C.; García, M.J.; León-Solís, L.E.; Rivera-Gutiérrez, S. Organization and Characterization of the Promoter Elements of the rRNA Operons in the Slow-Growing Pathogen Mycobacterium kumamotonense. Genes 2023, 14, 1023. https://doi.org/10.3390/genes14051023
Sánchez-Estrada R, Méndez-Guerrero O, García-Morales L, González-y-Merchand JA, Cerna-Cortes JF, Menendez MC, García MJ, León-Solís LE, Rivera-Gutiérrez S. Organization and Characterization of the Promoter Elements of the rRNA Operons in the Slow-Growing Pathogen Mycobacterium kumamotonense. Genes. 2023; 14(5):1023. https://doi.org/10.3390/genes14051023
Chicago/Turabian StyleSánchez-Estrada, Ricardo, Oscar Méndez-Guerrero, Lázaro García-Morales, Jorge Alberto González-y-Merchand, Jorge Francisco Cerna-Cortes, María Carmen Menendez, María Jesús García, Lizbel Esperanza León-Solís, and Sandra Rivera-Gutiérrez. 2023. "Organization and Characterization of the Promoter Elements of the rRNA Operons in the Slow-Growing Pathogen Mycobacterium kumamotonense" Genes 14, no. 5: 1023. https://doi.org/10.3390/genes14051023
APA StyleSánchez-Estrada, R., Méndez-Guerrero, O., García-Morales, L., González-y-Merchand, J. A., Cerna-Cortes, J. F., Menendez, M. C., García, M. J., León-Solís, L. E., & Rivera-Gutiérrez, S. (2023). Organization and Characterization of the Promoter Elements of the rRNA Operons in the Slow-Growing Pathogen Mycobacterium kumamotonense. Genes, 14(5), 1023. https://doi.org/10.3390/genes14051023





