Selection and Validation of Reference Genes for Gene Expression Studies in an Equine Adipose-Derived Mesenchymal Stem Cell Differentiation Model by Proteome Analysis and Reverse-Transcriptase Quantitative Real-Time PCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Preparation and Treatments
2.2. Cell Culture and Experimental Design
2.3. Cell Harvesting and Storage
2.4. Sample Preparation for LCMS3
2.5. Selection of Candidate RGs from Proteome Data
2.6. Primer Design
2.7. Determination of Optimal Annealing Temperatures for Selected Primers
2.8. Isolation of RNA and Synthesis of Complementary DNA
2.9. Quantitative Real-Time PCR
2.10. Analysis of TG Expression
2.11. Determination of Protein Fold Differences
3. Results
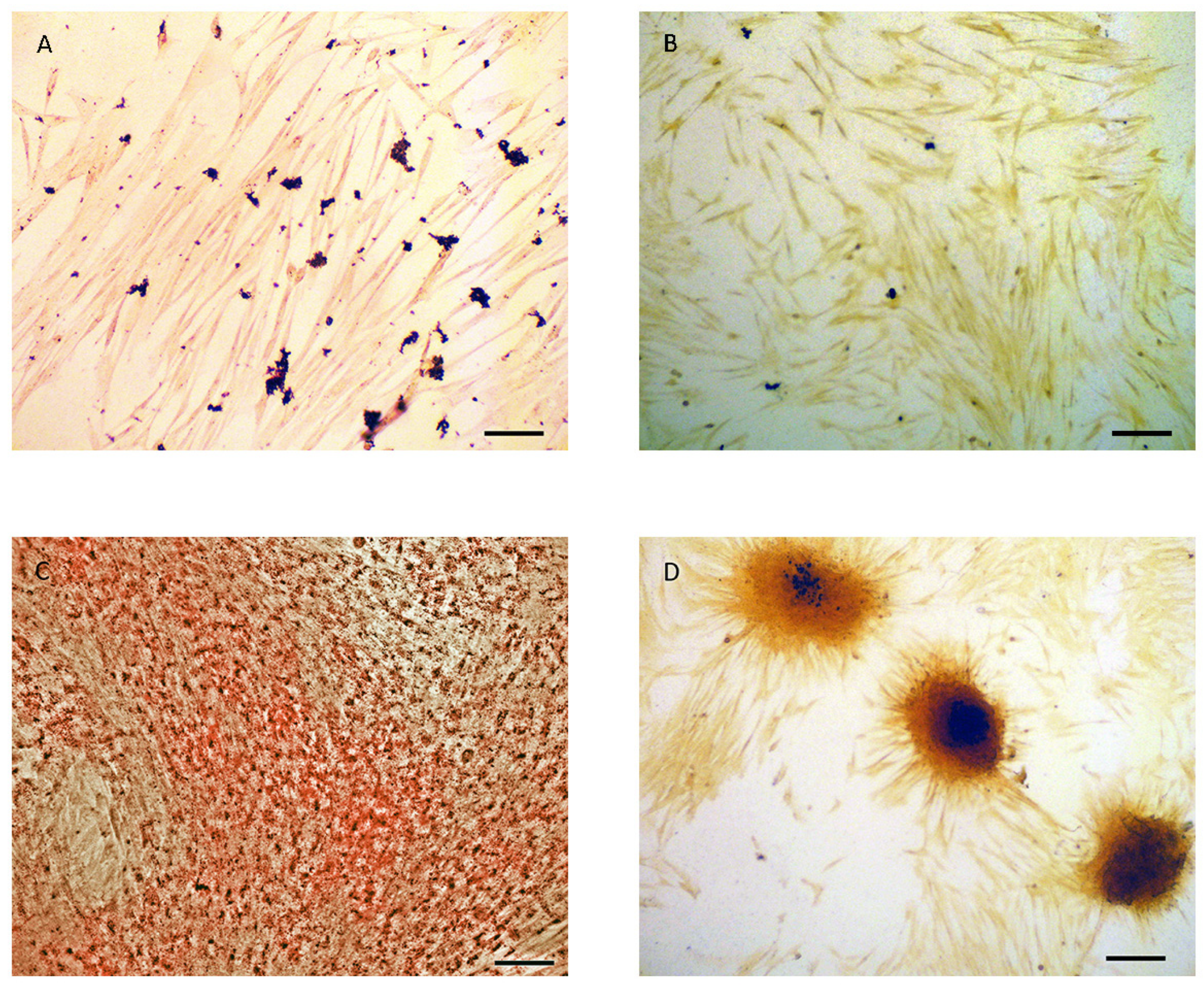
3.1. Validation of ADSCs Differentiation into ADs and OBs
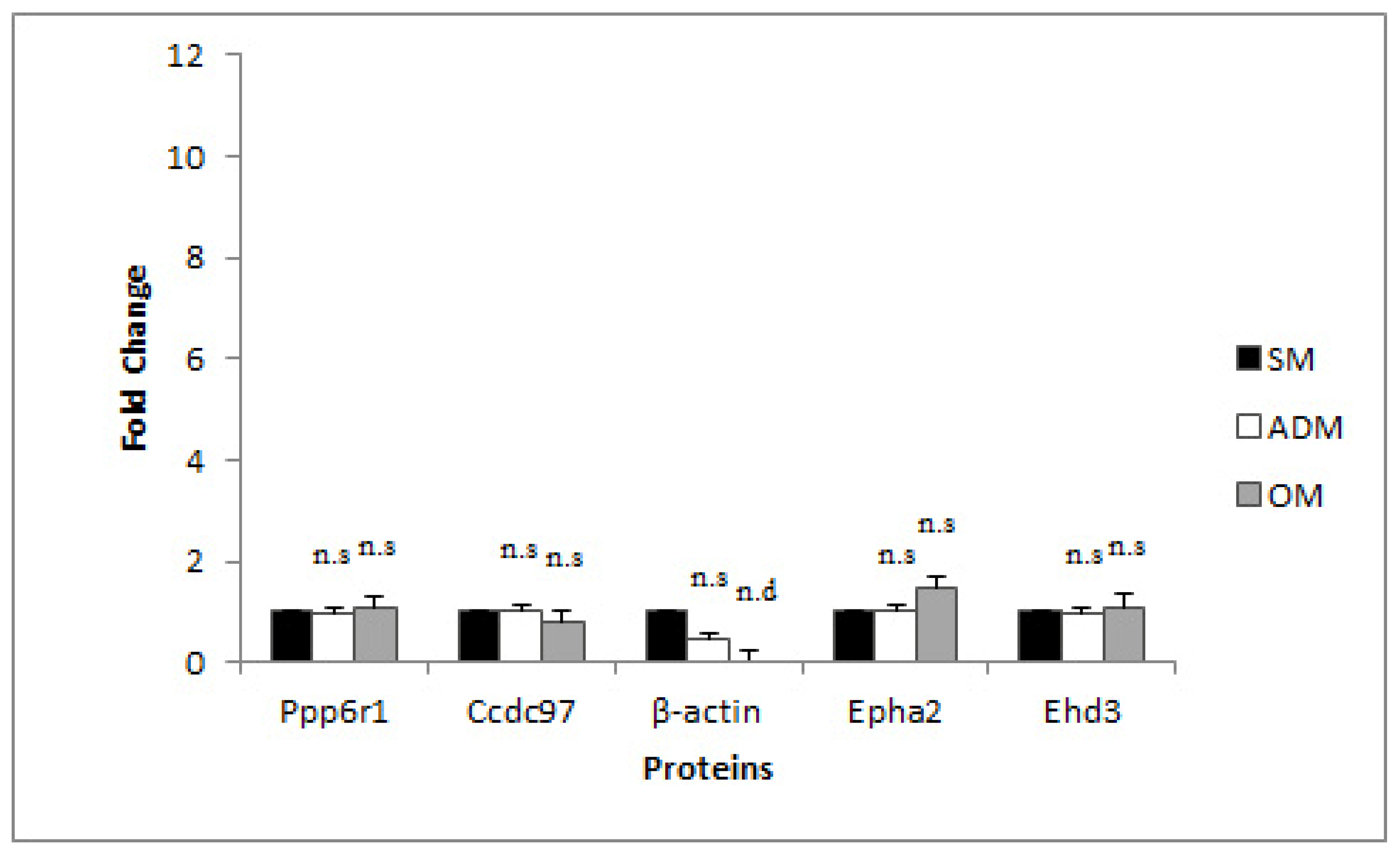
3.2. Candidate RG Selection for RT-qPCR
3.3. RG Stability Profiles
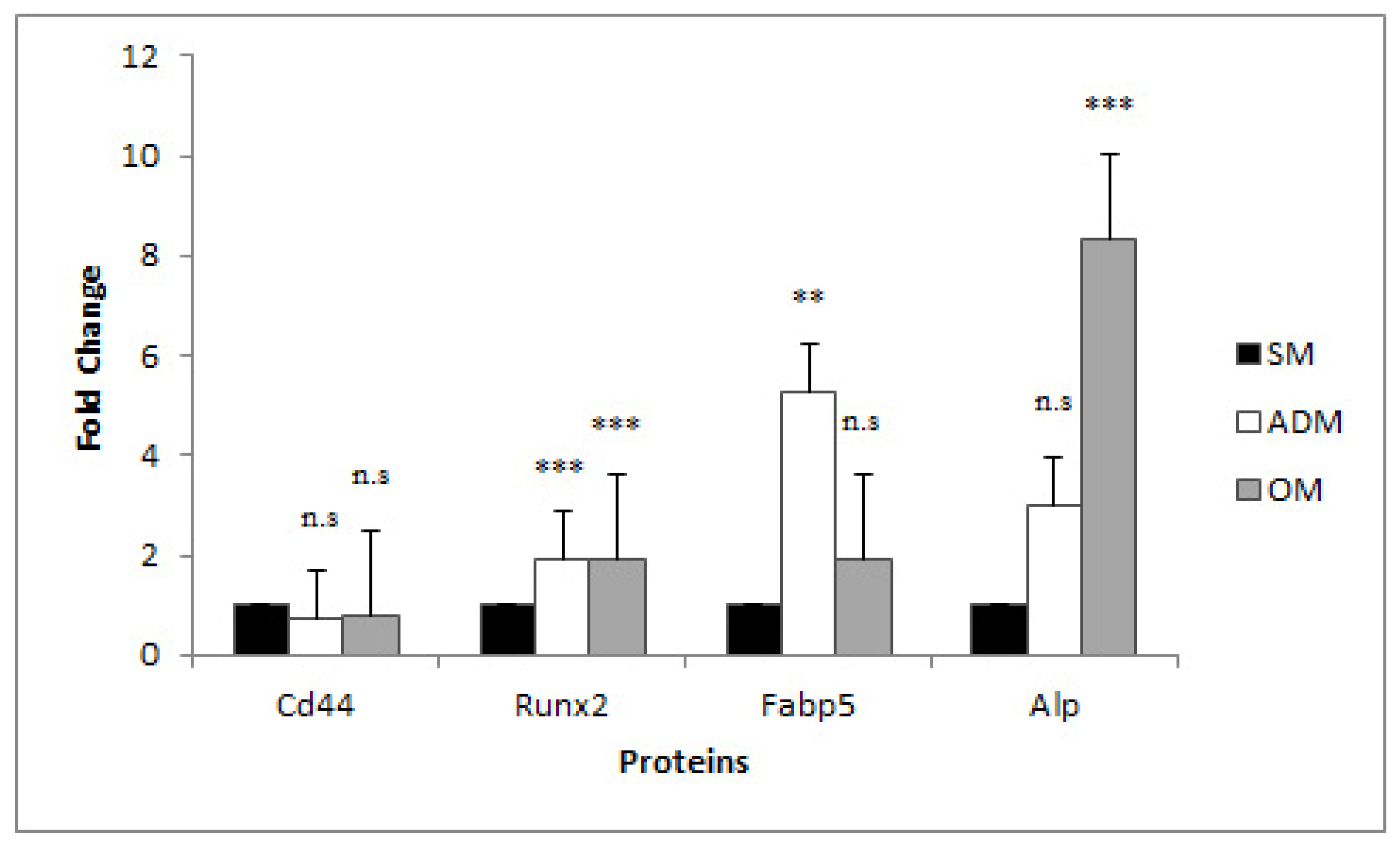
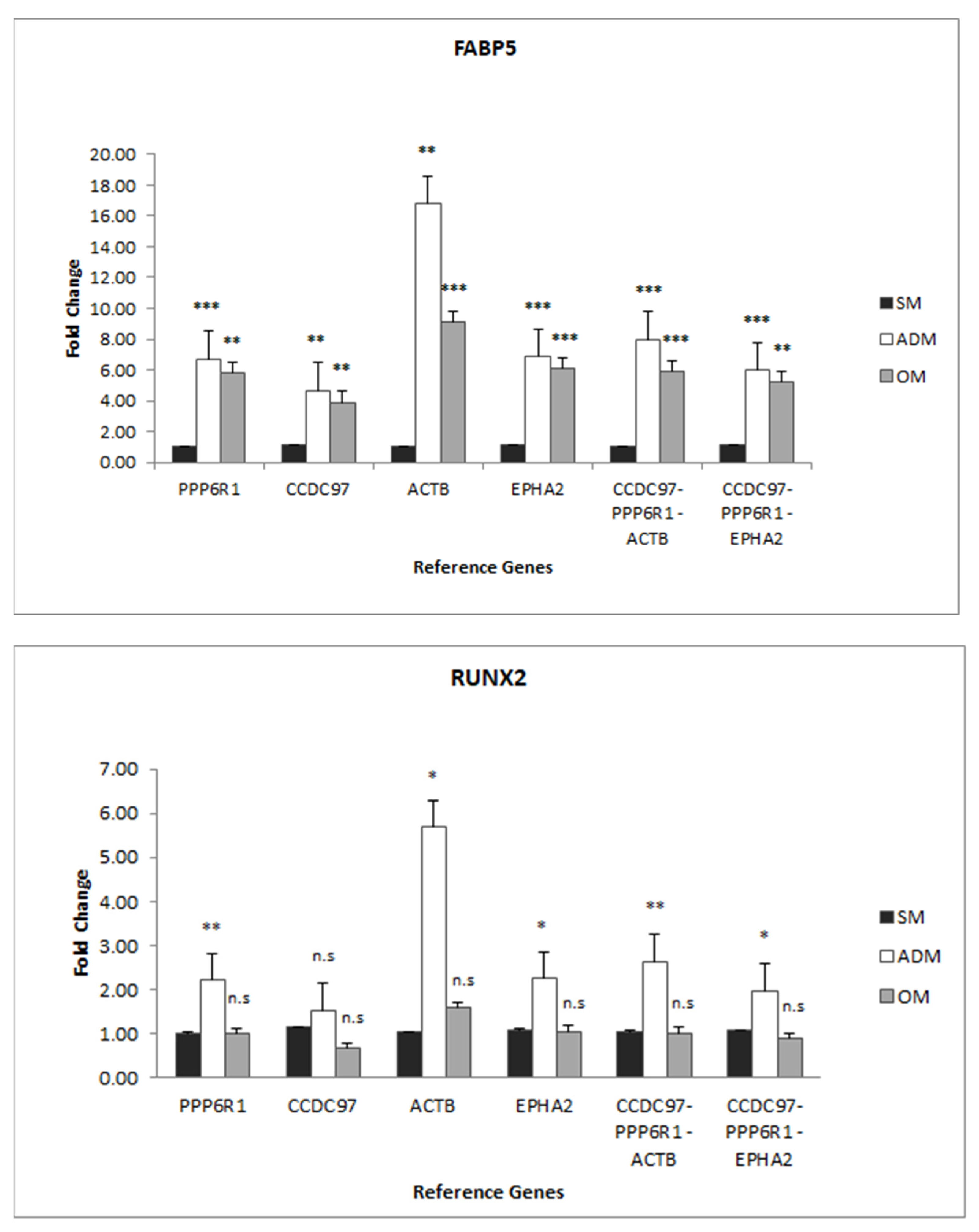
3.4. mRNA Expression profiles of TGs in ADSCs, Adipocytes, and Osteoblasts
4. Discussion
4.1. RG Congruence
4.2. TG Expression
4.3. The Impact of Primer Design and Experimental Model Specificity for RG Selection
5. Conclusions
Furture Studies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Radtke, C.L.; Nino-Fong, R.; Gonzalez, B.P.E.; Stryhn, H.; McDuffee, L.A. Characterization and osteogenic potential of equine muscle tissue- and periosteal tissue- derived mesenchymal stem cells in comparison with bone marrow- and adipose tissue- derived mesenchymal stem cells. Am. J. Vet. Res. 2013, 74, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Ranera, B.; Ordovás, L.; Lyahyai, J.; Bernal, M.L.; Fernandes, F.; Remacha, A.R.; Romero, A.; Vázquez, F.J.; Osta, R.; Cons, C.; et al. Comparative study of equine bone marrow and adipose tissue-derived mesenchymal stromal cells. Equine Vet. J. 2012, 44, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Davis, E.G.; Bai, J.F. Determination of internal control for gene expression studies in equine tissues and cell culture using quantitative RT-PCR. Vet. Immunol. Immunopathol. 2009, 130, 114–119. [Google Scholar] [CrossRef]
- Ranera, B.; Lyahyai, J.; Romero, A.; Vázquez, F.J.; Remacha, A.R.; Bernal, M.L.; Zaragoza, P.; Rodellar, C.; Martín-Burriel, I. Immunophenotype and gene expression profiles of cell surface markers of mesenchymal stem cells derived from equine bone marrow and adipose tissue. Vet. Immunol. Immunopathol. 2011, 144, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Nazari, F.; Parham, A.; Maleki, A.F. GAPDH, β-actin and β2-microglobulin, as three common reference genes, are not reliable for gene expression studies in equine adipose- and marrow-derived mesenchymal stem cells. J. Anim. Sci. Technol. 2015, 57, 1–8. [Google Scholar] [CrossRef]
- Busato, S.; Mezzetti, M.; Logan, P.; Aguilera, N.; Bionaz, M. What’s the norm in normalization? A frightening note on the use of RT-qPCR in the livestock science. Gene 2019, 721, 100003. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated TGs and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Bunnell, B.A.; Estes, B.T.; Guilak, F.; Gimble, J.M. Differentiation of adipose stem cells. Methods Mol. Biol. 2008, 456, 155–171. [Google Scholar] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Bradburn, S. How to Analyse qPCR Results with Multiple Reference Genes [Internet]. Top Tip Bio. 2018. Available online: https://toptipbio.com/qpcr-multiple-reference-genes/ (accessed on 4 September 2021).
- Bradburn, S. How to Perform the Pfaffl Method for qPCR [Internet]. Top Tip Bio. 2018. Available online: https://toptipbio.com/pfaffl-method-qpcr/ (accessed on 4 September 2021).
- Yamamoto, T.; Furuhashi, M.; Sugaya, T.; Oikawa, T.; Matsumoto, M.; Funahashi, Y.; Matsukawa, Y.; Gotoh, M.; Miura, T. Transcriptome and Metabolome Analyses in Exogenous FABP4- and FABP5-Treated Adipose-Derived Stem Cells. PLoS ONE 2016, 11, e0167825. [Google Scholar] [CrossRef]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Komori, T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Li, X.; Qian, S.-W.; Guo, L.; Huang, H.-Y.; He, Q.; Liu, Y.; Ma, C.-G.; Tang, Q.-Q. Down-Regulation of Type I Runx2 Mediated by Dexamethasone Is Required for 3T3-L1 Adipogenesis. Mol. Endocrinol. 2012, 26, 798–808. [Google Scholar] [CrossRef]
- Vedula, P.; Kurosaka, S.; MacTaggart, B.; Ni, Q.; Papoian, G.; Jiang, Y.; Dong, D.W.; Kashina, A. Different translation dynamics of β- and γ-actin regulates cell migration. eLife 2021, 10, e68712. [Google Scholar] [CrossRef] [PubMed]
- Abd-El-Aziz, A.; Riveroll, A.; Esparza-Gonsalez, B.; McDuffee, L.; Cohen, A.M.; Fenech, A.L.; Montelpare, W.J. Heat Shock Alters the Proteomic Profile of Equine Mesenchymal Stem Cells. Int. J. Mol. Sci. 2022, 23, 7233. [Google Scholar] [CrossRef] [PubMed]
| Sample No. | Condition | Reagent |
|---|---|---|
| S1 | SM1 | TMT10-126 |
| S2 | SM2 | TMT10-127N |
| S3 | SM3 | TMT10-127C |
| S4 | OM1 | TMT10-128N |
| S5 | OM2 | TMT10-128C |
| S6 | OM3 | TMT10-129N |
| S7 | AD1 | TMT10-129C |
| S8 | AD2 | TMT10-130N |
| S9 | AD3 | TMT10-130C |
| Sample Pool 1 | SM1–AD3 | TMT10-131 |
| Ranking | Uniprot Accession Number (GN = Corresponding Gene Name) | Protein Description | Difference of Means ADSC-AD (MRPA) | Difference of Means ADSC-OB (MRPA) | Difference of Means AD-OB (MRPA) | ANOVA (p-Value) * |
|---|---|---|---|---|---|---|
| 1 | F6RYC9 GN = FMC1 | Formation of mitochondrial complex V assembly factor 1 homolog | 0.09 | 203.33 | 203.25 | 0.71 |
| 2 | F6VEQ6 GN = BCKDK | Branched chain ketoacid dehydrogenase kinase | 1.41 | 841.22 | 839.81 | 0.61 |
| 3 | F6S466 GN = n.r. | Uncharacterized protein | 2.45 | 362.92 | 360.47 | 0.10 |
| 4 | F7C673 GN = ARHGAP12 | Rho GTPase activating protein 12 | 2.66 | 25.72 | 28.38 | 0.99 |
| 5 | F7DQ27 GN = GLMN | Glomulin, FKBP associated protein | 2.81 | 439.74 | 436.94 | 0.63 |
| 6 | F7DY57 GN = SLC12A5 | Solute carrier family 12 member 5 | 3.41 | 152.02 | 148.61 | 0.64 |
| 7 | F6QJT2 GN = TWSG1 | Twisted gastrulation BMP signaling modulator 1 | 6.43 | 122.23 | 115.80 | 0.99 |
| 8 | F7CVS4 GN = EHD3 | EH domain containing 3 | 6.89 | 76.78 | 83.66 | 0.91 |
| 9 | F6XYF8 GN = TRIM26 | Tripartite motif containing 26 | 7.04 | 506.65 | 499.61 | 0.46 |
| 10 | F6SKN5 GN = LRSAM1 | Leucine rich repeat and sterile α motif containing 1 | 7.34 | 77.12 | 69.78 | 0.85 |
| 11 | F6PTR6 GN = PPP1R9B | Protein phosphatase 1 regulatory subunit 9B | 7.50 | 3534.30 | 3526.79 | 0.37 |
| 12 | F7D5F3 GN = SDHC | Succinate dehydrogenase complex subunit C | 7.69 | 159.14 | 166.82 | 0.82 |
| 13 | F7A2B6 GN = CHM | Rab proteins geranylgeranyltransferase component A | 7.71 | 339.83 | 347.55 | 0.77 |
| 14 | F6QAL5 GN = EPHA2 | EPH receptor A2 | 8.22 | 1153.61 | 1161.84 | 0.73 |
| 15 | F6SG13 GN = MPHOSPH8 | Uncharacterized protein | 9.24 | 51.53 | 60.77 | 0.96 |
| 16 | F6R625 GN = CCDC97 | Coiled-coil domain containing 97 | 9.25 | 159.47 | 168.72 | 0.36 |
| 17 | F6TZB2 GN = NRBP1 | Nuclear receptor binding protein 1 | 9.43 | 64.97 | 74.40 | 0.99 |
| 18 | H9H022 GN = n.r. | PHD finger protein 8 | 9.46 | 50.95 | 60.41 | 0.70 |
| 19 | F6V860 GN = PPP6R1 | Protein phosphatase 6 regulatory subunit 1 | 9.78 | 27.80 | 37.58 | 0.86 |
| 20 | F6PMV9 GN = IRGQ | Immunity related GTPase Q | 10.82 | 278.48 | 267.66 | 0.79 |
| 21 | F6WE69 GN = ARHGEF12 | Rho guanine nucleotide exchange factor 12 | 11.16 | 86.06 | 74.90 | 0.95 |
| 22 | F6T4G5 GN = DCAF8 | Uncharacterized protein | 12.01 | 883.61 | 895.62 | 0.77 |
| 23 | F7DNN2 GN = COQ4 | Ubiquinone biosynthesis protein COQ4 homolog, mitochondrial | 12.08 | 488.65 | 476.57 | 0.23 |
| Genes (NCBI Record) | Forward and Reverse Primer Sequences (5′ > 3′) | Amplicon Size (bp) | Annealing Temperature for qPCR (°C) | R2 | E (%) | Mean Cq ± STDEV | Intron Splice Site (Nucleotide Position Based on NCBI Record) |
|---|---|---|---|---|---|---|---|
| PPP6R1 (XM_014730802.2) | F840 ATTGTCCAGCGGCTCATCGAGC R912 GGGACTGGGACGCATTGGAATG | 73 | 67 | 0.9535 | 88.3 | 24.50 ± 0.2 | 887, 888 |
| EHD3 (XM_001918104.5) | F1121 CTTTGGCAATGCCTTCTTGAAC R1217 AGAGAGGATCCCTGGAGTGTCG | 97 | 62 | 0.9954 | 107.4 | 24.51 ± 0.08 | 1144, 1145 |
| CCDC97 (XM_001500237.6) | F501 TGGTGACCACCGAGCAGACTTC R647 TGCTCGTCGCTGAAGTACTCGC | 147 | 71 | 0.9941 | 109.92 | 23.67 ± 0.04 | 622, 623 |
| EPHA2 (XM_001488739.5) | F764 TGCCCATCGGTCAGTGTCTGTG R888 GTGTGCAGGGCACTCCAAACAG | 125 | 71 | 0.9842 | 101.61 | 20.84 ± 0.14 | 823, 824 |
| GAPDH (NM_001163856.1) | F552 TGGCATCGTGGAGGGACTCATG R642 ATCGCGCCACATCTTCCCAGAG | 91 | 72 | 0.9783 | 90.98 | 17.84 ± 0.23 | 573, 574 |
| ACTB (NM_001081838.1) | F924 CATCGCCGACAGGATGCAGAAG R1060 GCTGGAAGGTGGACAATGAGGC | 137 | 72 | 0.9329 | 109.42 | 16.19 ± 0.35 | 984, 985 |
| β2M (NM_001082502.3) | F14 CTGCTGCTGTGGTAGCTATGGC R118 AAACCTGAACCTTCGGAACACG | 105 | 65 | 0.9479 | 83.65 | 21.19 ± 0.24 | 97, 98 |
| FABP5 (XM_001489456.5) | F103 GAAGATGGCGCTTGGTGGAGAG R204 AATCTGGTTTGGCCATTGCACC | 102 | 68 | 0.9723 | 103.98 | 26.75 ± 0.37 | 156, 157 |
| RUNX2 (XM_023624251.1) | F847 CTGCTGAGCTCCGAAATGCCTC R942 AACTCTTGCCTCGTCCACTCCG | 96 | 69 | 0.9348 | 90.2 | 23.89 ± 0.14 | 934, 935 |
| Rank | BestKeeper | NormFinder | geNorm | ∆CT analysis | RefFinder Analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | r * (STDEV, p-value) | Gene | Stability Value (SE) | Gene | M Value | Gene | Average of STDEV | Gene | Geometric Mean of Ranking Values | |
| 1 | CCDC97 | 0.943 (0.29, 0.001) | PPP6R1 | 0.005 (0.007) | PPP6R1 | 0.312 | PPP6R1 | 0.42 | PPP6R1 | 1.19 |
| 2 | PPP6R1 | 0.883 (0.19, 0.002) | CCDC97 | 0.005 (0.007) | CCDC97 | 0.329 | CCDC97 | 0.46 | CCDC97 | 1.86 |
| 3 | GAPDH | 0.769 (0.58, 0.015) | ACTB | 0.010 (0.010) | EPHA2 | 0.354 | ACTB | 0.47 | ACTB | 2.28 |
| 4 | EPHA2 | 0.463 (0.35, 0.210) | EPHA2 | 0.019 (0.006) | GAPDH | 0.376 | EPHA2 | 0.58 | EPHA2 | 4.00 |
| 5 | β2M | 0.309 (0.44, 0.418) | EHD3 | 0.026 (0.007) | ACTB | 0.427 | β2M | 0.62 | β2M | 5.48 |
| 6 | ACTB | 0.058 (0.06, 0.885) | β2M | 0.030 (0.008) | β2M | 0.526 | GAPDH | 0.66 | GAPDH | 5.96 |
| 7 | EHD3 | −0.057 (0.44, 0.885) | GAPDH | 0.040 (0.010) | EHD3 | 0.601 | EHD3 | 0.68 | EHD3 | 6.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riveroll, A.L.; Skyba-Lewin, S.; Lynn, K.D.; Mubyeyi, G.; Abd-El-Aziz, A.; Kibenge, F.S.T.; Kibenge, M.J.T.; Cohen, A.M.; Esparza-Gonsalez, B.; McDuffee, L.; et al. Selection and Validation of Reference Genes for Gene Expression Studies in an Equine Adipose-Derived Mesenchymal Stem Cell Differentiation Model by Proteome Analysis and Reverse-Transcriptase Quantitative Real-Time PCR. Genes 2023, 14, 673. https://doi.org/10.3390/genes14030673
Riveroll AL, Skyba-Lewin S, Lynn KD, Mubyeyi G, Abd-El-Aziz A, Kibenge FST, Kibenge MJT, Cohen AM, Esparza-Gonsalez B, McDuffee L, et al. Selection and Validation of Reference Genes for Gene Expression Studies in an Equine Adipose-Derived Mesenchymal Stem Cell Differentiation Model by Proteome Analysis and Reverse-Transcriptase Quantitative Real-Time PCR. Genes. 2023; 14(3):673. https://doi.org/10.3390/genes14030673
Chicago/Turabian StyleRiveroll, Angela L., Sabrina Skyba-Lewin, K. Devon Lynn, Glady’s Mubyeyi, Ahmad Abd-El-Aziz, Frederick S. T. Kibenge, Molly J. T. Kibenge, Alejandro M. Cohen, Blanca Esparza-Gonsalez, Laurie McDuffee, and et al. 2023. "Selection and Validation of Reference Genes for Gene Expression Studies in an Equine Adipose-Derived Mesenchymal Stem Cell Differentiation Model by Proteome Analysis and Reverse-Transcriptase Quantitative Real-Time PCR" Genes 14, no. 3: 673. https://doi.org/10.3390/genes14030673
APA StyleRiveroll, A. L., Skyba-Lewin, S., Lynn, K. D., Mubyeyi, G., Abd-El-Aziz, A., Kibenge, F. S. T., Kibenge, M. J. T., Cohen, A. M., Esparza-Gonsalez, B., McDuffee, L., & Montelpare, W. J. (2023). Selection and Validation of Reference Genes for Gene Expression Studies in an Equine Adipose-Derived Mesenchymal Stem Cell Differentiation Model by Proteome Analysis and Reverse-Transcriptase Quantitative Real-Time PCR. Genes, 14(3), 673. https://doi.org/10.3390/genes14030673







